You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000013_02447
You are here: Home > Sequence: MGYG000000013_02447
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
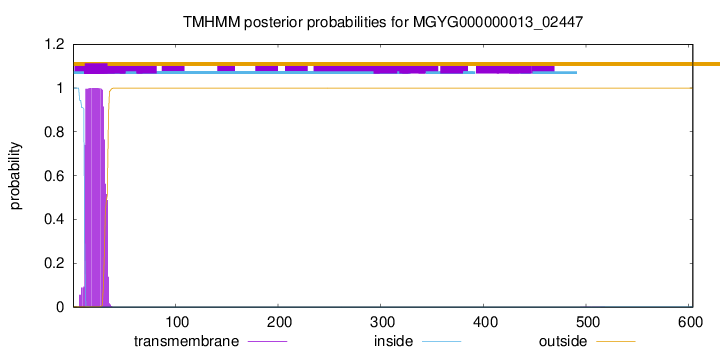
TMHMM annotations
Basic Information help
| Species | Bacteroides sp902362375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp902362375 | |||||||||||
| CAZyme ID | MGYG000000013_02447 | |||||||||||
| CAZy Family | CBM85 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 472423; End: 474237 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 253 | 535 | 1.5e-66 | 0.9372937293729373 |
| CBM85 | 56 | 185 | 3.2e-22 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.67e-44 | 311 | 534 | 20 | 258 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.40e-41 | 256 | 534 | 13 | 303 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 7.85e-32 | 280 | 530 | 55 | 318 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEE54613.1 | 5.27e-158 | 28 | 418 | 29 | 420 |
| CCW34445.1 | 3.21e-118 | 59 | 588 | 54 | 578 |
| QDU72038.1 | 4.30e-113 | 91 | 598 | 104 | 605 |
| AHF89713.1 | 8.38e-112 | 93 | 580 | 128 | 616 |
| QYM77625.1 | 3.96e-109 | 30 | 582 | 23 | 583 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 3.94e-54 | 208 | 586 | 41 | 409 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 7.55e-53 | 208 | 586 | 41 | 409 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1NQ6_A | 6.14e-25 | 255 | 534 | 15 | 293 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 1VBR_A | 1.10e-23 | 271 | 546 | 30 | 323 | Crystalstructure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
| 3NIY_A | 1.35e-23 | 271 | 546 | 46 | 339 | Crystalstructure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 6.32e-48 | 211 | 591 | 380 | 749 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| A0A1P8AWH8 | 3.56e-32 | 213 | 574 | 562 | 915 | Endo-1,4-beta-xylanase 1 OS=Arabidopsis thaliana OX=3702 GN=XYN1 PE=1 SV=1 |
| Q84WT5 | 3.41e-31 | 211 | 547 | 173 | 505 | Endo-1,4-beta-xylanase 5-like OS=Arabidopsis thaliana OX=3702 GN=At4g33820 PE=2 SV=1 |
| O80596 | 9.07e-29 | 239 | 595 | 710 | 1057 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| A0A1P8B8F8 | 2.48e-23 | 211 | 576 | 173 | 539 | Endo-1,4-beta-xylanase 5 OS=Arabidopsis thaliana OX=3702 GN=XYN5 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
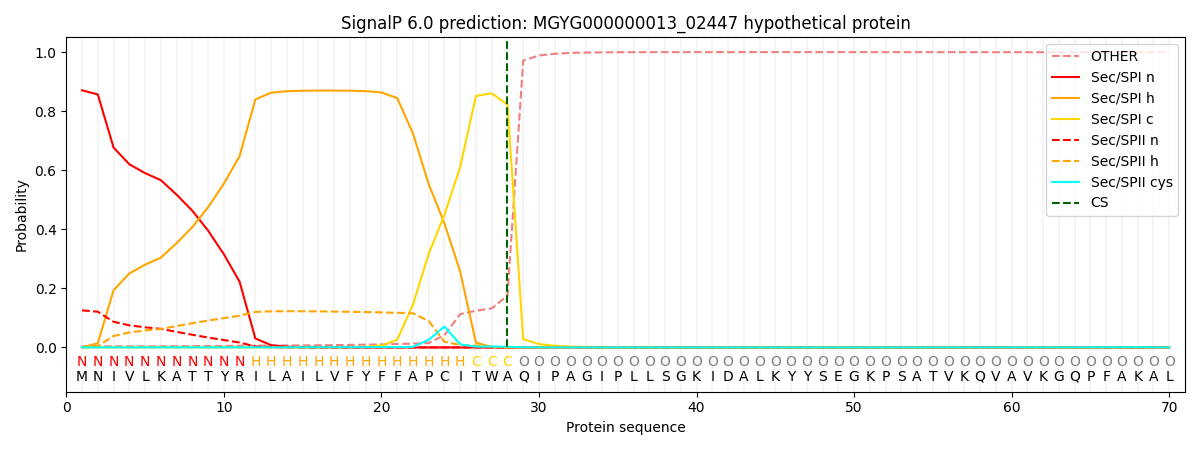
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005484 | 0.864016 | 0.129723 | 0.000283 | 0.000236 | 0.000220 |