You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000043_00538
You are here: Home > Sequence: MGYG000000043_00538
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900066455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900066455 | |||||||||||
| CAZyme ID | MGYG000000043_00538 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47172; End: 48863 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM67 | 353 | 527 | 7.4e-33 | 0.8977272727272727 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08531 | Bac_rhamnosid_N | 3.38e-57 | 371 | 528 | 2 | 155 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VDS02406.1 | 3.78e-310 | 25 | 563 | 8 | 546 |
| VDS02598.1 | 5.86e-310 | 28 | 563 | 3 | 538 |
| VDS02663.1 | 5.86e-310 | 28 | 563 | 3 | 538 |
| VDS02632.1 | 5.86e-310 | 28 | 563 | 3 | 538 |
| QUT88780.1 | 4.08e-217 | 27 | 562 | 54 | 593 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W5M_A | 2.32e-52 | 32 | 537 | 10 | 478 | CrystalStructure of Streptomyces avermitilis alpha-L-rhamnosidase [Streptomyces avermitilis MA-4680 = NBRC 14893],3W5N_A Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose [Streptomyces avermitilis MA-4680 = NBRC 14893] |
| 6I60_A | 2.41e-29 | 359 | 524 | 177 | 328 | Structureof alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12],6I60_B Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum [Dictyoglomus thermophilum H-6-12] |
| 6GSZ_A | 5.31e-28 | 362 | 559 | 135 | 329 | Crystalstructure of native alfa-L-rhamnosidase from Aspergillus terreus [Aspergillus terreus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q82PP4 | 1.23e-51 | 32 | 537 | 10 | 478 | Alpha-L-rhamnosidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=SAVERM_828 PE=1 SV=1 |
| T2KNB2 | 6.88e-24 | 369 | 521 | 184 | 332 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22090 PE=1 SV=2 |
| P9WF03 | 1.61e-23 | 361 | 520 | 173 | 327 | Alpha-L-rhamnosidase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_34 PE=1 SV=1 |
| T2KPL4 | 1.74e-18 | 369 | 546 | 194 | 371 | Alpha-L-rhamnosidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22170 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
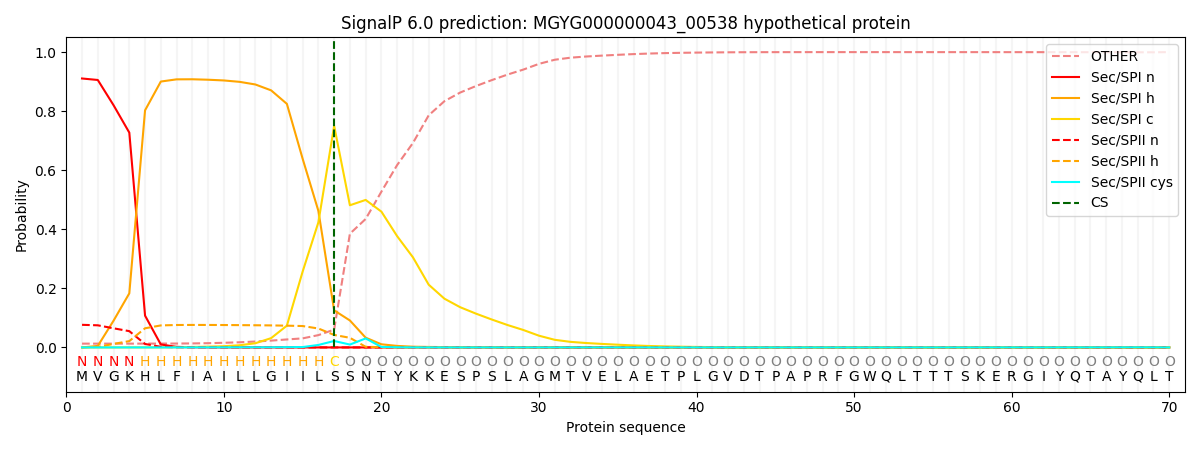
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014851 | 0.905117 | 0.079215 | 0.000286 | 0.000261 | 0.000243 |
