You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000057_01370
You are here: Home > Sequence: MGYG000000057_01370
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp002491635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp002491635 | |||||||||||
| CAZyme ID | MGYG000000057_01370 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61326; End: 63023 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 359 | 560 | 1.1e-63 | 0.6567656765676567 |
| GH26 | 143 | 291 | 7e-24 | 0.44224422442244227 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.87e-30 | 287 | 533 | 22 | 288 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 7.51e-18 | 406 | 529 | 180 | 319 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam13205 | Big_5 | 3.12e-04 | 31 | 132 | 1 | 106 | Bacterial Ig-like domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU00593.1 | 0.0 | 1 | 565 | 1 | 565 |
| BBK85892.1 | 0.0 | 1 | 565 | 1 | 565 |
| QUT62087.1 | 0.0 | 1 | 565 | 1 | 565 |
| QUT33706.1 | 0.0 | 1 | 565 | 1 | 565 |
| QQA30553.1 | 0.0 | 1 | 565 | 1 | 565 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6HF2_A | 7.87e-91 | 122 | 563 | 29 | 358 | Thestructure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus [Bacteroides ovatus ATCC 8483],6HF4_A The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 [Bacteroides ovatus ATCC 8483] |
| 6D2X_A | 4.24e-76 | 141 | 561 | 7 | 334 | Crystalstructure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii [Prevotella bryantii B14] |
| 6D2W_A | 1.13e-71 | 141 | 561 | 98 | 425 | Crystalstructure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14],6D2W_B Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14] |
| 3WDQ_A | 1.10e-40 | 357 | 562 | 138 | 353 | Crystalstructure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus [Symbiotic protist of Reticulitermes speratus],3WDR_A Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide [Symbiotic protist of Reticulitermes speratus] |
| 3ZM8_A | 4.04e-27 | 324 | 529 | 215 | 418 | ChainA, Gh26 Endo-beta-1,4-mannanase [Podospora anserina S mat+] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49425 | 3.23e-35 | 357 | 562 | 250 | 458 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
| P55298 | 6.37e-30 | 361 | 563 | 263 | 461 | Mannan endo-1,4-beta-mannosidase C OS=Piromyces sp. OX=45796 GN=MANC PE=2 SV=1 |
| P55297 | 6.46e-30 | 361 | 563 | 265 | 463 | Mannan endo-1,4-beta-mannosidase B OS=Piromyces sp. OX=45796 GN=MANB PE=2 SV=1 |
| P55296 | 6.71e-28 | 361 | 563 | 264 | 462 | Mannan endo-1,4-beta-mannosidase A OS=Piromyces sp. OX=45796 GN=MANA PE=2 SV=1 |
| Q5AWB7 | 2.44e-25 | 299 | 527 | 69 | 319 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
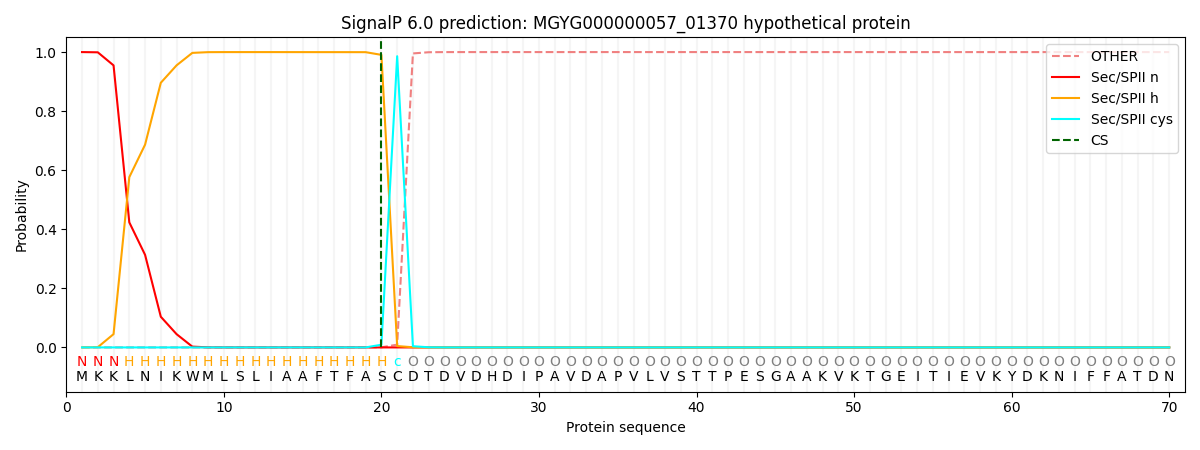
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000035 | 0.000000 | 0.000000 | 0.000000 |
