You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000070_00298
You are here: Home > Sequence: MGYG000000070_00298
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
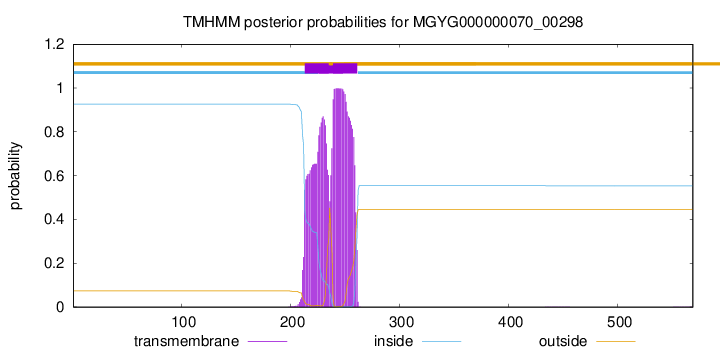
TMHMM annotations
Basic Information help
| Species | Lawsonibacter sp900066825 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Lawsonibacter; Lawsonibacter sp900066825 | |||||||||||
| CAZyme ID | MGYG000000070_00298 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 301121; End: 302830 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05257 | CHAP | 1.25e-15 | 454 | 544 | 1 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
| TIGR02594 | TIGR02594 | 0.002 | 451 | 564 | 27 | 127 | TIGR02594 family protein. Members of this protein family known so far are restricted to the bacteria, and for the most to the proteobacteria. The function is unknown. |
| TIGR00984 | 3a0801s03tim44 | 0.009 | 84 | 166 | 14 | 108 | mitochondrial import inner membrane, translocase subunit. The mitochondrial protein translocase (MPT) family, which brings nuclearly encoded preproteins into mitochondria, is very complex with 19 currently identified protein constituents.These proteins include several chaperone proteins, four proteins of the outer membrane translocase (Tom) import receptor, five proteins of the Tom channel complex, five proteins of the inner membrane translocase (Tim) and three "motor" proteins. This family is specific for the Tim proteins. [Transport and binding proteins, Amino acids, peptides and amines] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYH40048.1 | 1.07e-112 | 1 | 569 | 1 | 567 |
| CBL27479.1 | 3.00e-112 | 1 | 569 | 1 | 567 |
| QOV19260.1 | 2.35e-111 | 1 | 569 | 1 | 567 |
| QJA01756.1 | 1.26e-98 | 1 | 569 | 1 | 587 |
| AVM68330.1 | 3.82e-97 | 1 | 569 | 1 | 587 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
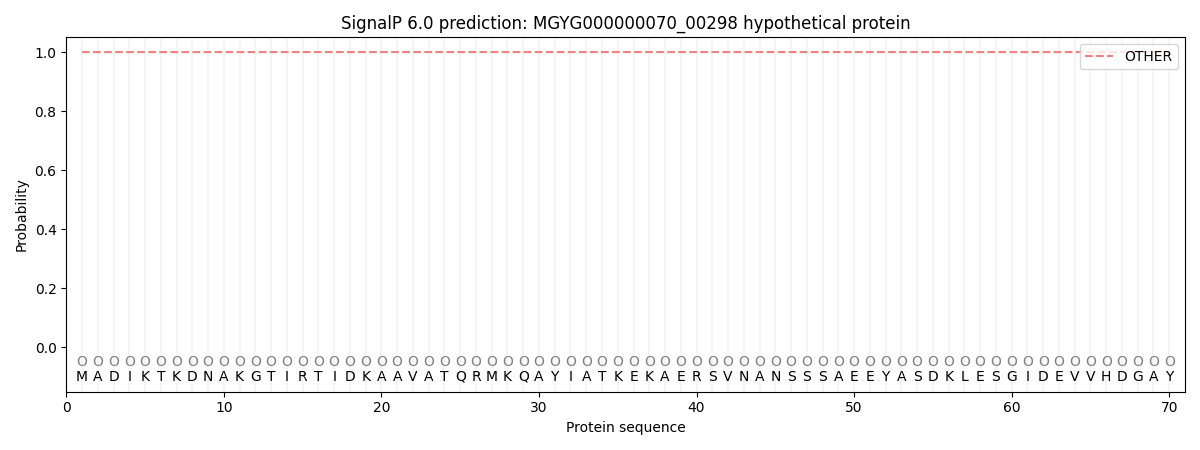
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000058 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |