You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000089_01338
You are here: Home > Sequence: MGYG000000089_01338
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-353 sp900066885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; CAG-353; CAG-353 sp900066885 | |||||||||||
| CAZyme ID | MGYG000000089_01338 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 254240; End: 257329 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 78 | 352 | 2e-86 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 5.63e-54 | 73 | 352 | 10 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.33e-30 | 57 | 358 | 45 | 366 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam18259 | CBM65_1 | 5.30e-06 | 401 | 502 | 1 | 113 | Carbohydrate binding module 65 domain 1. This domain is found in the non-catalytic carbohydrate binding module 65B (CMB65B) present in Eubacterium cellulosolvens. CBMs are present in plant cell wall degrading enzymes and are responsible for targeting, which enhances catalysis. CBM65s display higher affinity for oligosaccharides, such as cellohexaose, and particularly polysaccharides than cellotetraose, which fully occupies the core component of the substrate binding cleft. The concave surface presented by beta-sheet 2 comprises the beta-glucan binding site in CBM65s. C6 of all the backbone glucose moieties makes extensive hydrophobic interactions with the surface tryptophans of CBM65s. Three out of the four surface Trp are highly conserved. The conserved metal ion site typical of CBMs is absent in this CBM65 family. |
| COG3401 | FN3 | 0.002 | 670 | 944 | 9 | 295 | Fibronectin type 3 domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEI34646.1 | 5.96e-123 | 20 | 503 | 344 | 845 |
| AWV80935.1 | 5.96e-123 | 20 | 503 | 344 | 845 |
| AAK81397.1 | 5.96e-123 | 20 | 503 | 344 | 845 |
| ADZ22510.1 | 5.96e-123 | 20 | 503 | 344 | 845 |
| CUH91691.1 | 5.52e-119 | 32 | 503 | 329 | 809 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UI3_A | 9.47e-97 | 45 | 385 | 5 | 340 | GH5-4broad specificity endoglucanase from Clostridum cellulovorans [Clostridium cellulovorans] |
| 6WQP_A | 1.06e-79 | 48 | 381 | 8 | 353 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6MQ4_A | 6.25e-75 | 49 | 381 | 5 | 346 | ChainA, cellulase [Acetivibrio cellulolyticus] |
| 6PZ7_A | 9.53e-75 | 54 | 381 | 7 | 334 | GH5-4broad specificity endoglucanase from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 3NDY_A | 2.46e-74 | 49 | 386 | 5 | 343 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_A The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_B The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_C The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_D The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23660 | 1.25e-73 | 50 | 381 | 21 | 360 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28623 | 8.51e-73 | 1 | 386 | 1 | 374 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P54937 | 8.36e-72 | 55 | 384 | 38 | 378 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P28621 | 2.72e-71 | 49 | 366 | 35 | 356 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P16216 | 1.31e-65 | 55 | 388 | 68 | 402 | Endoglucanase 1 OS=Ruminococcus albus OX=1264 GN=Eg I PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
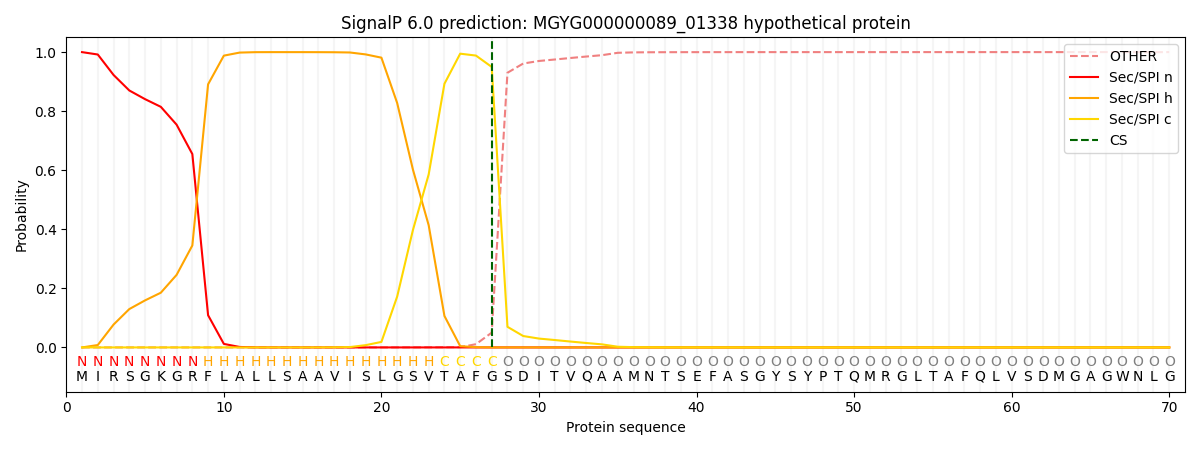
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000488 | 0.998623 | 0.000195 | 0.000271 | 0.000201 | 0.000181 |
