You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000105_01836
You are here: Home > Sequence: MGYG000000105_01836
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides clarus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides clarus | |||||||||||
| CAZyme ID | MGYG000000105_01836 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 405593; End: 408043 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 461 | 693 | 4.7e-31 | 0.7222222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13149 | Mfa_like_1 | 1.92e-36 | 46 | 301 | 1 | 244 | Fimbrillin-like. A family of putative fimbrillin proteins found by clustering human gut metagenomic sequences. Analysis of structural comparisons shows this family to be part of the FimbA (CL0450) superfamily of adhesin components or fimbrillins. |
| cd13120 | BF2867_like_N | 4.56e-17 | 10 | 167 | 1 | 156 | N-terminal domain found in Bacteroides fragilis Nctc 9343 BF2867 and related proteins. Two structurally similar domains with low sequence similarity in a tandem repeat arrangement form a protein that may have a role in cell adhesion. This family overlaps with DUF3988. |
| PLN02682 | PLN02682 | 1.11e-16 | 457 | 683 | 70 | 278 | pectinesterase family protein |
| PLN02432 | PLN02432 | 6.39e-15 | 590 | 683 | 109 | 203 | putative pectinesterase |
| PLN02304 | PLN02304 | 2.25e-13 | 450 | 682 | 67 | 284 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNL41098.1 | 1.37e-164 | 3 | 814 | 4 | 802 |
| QUT78059.1 | 3.83e-164 | 3 | 814 | 4 | 802 |
| SCV08390.1 | 3.83e-164 | 3 | 814 | 4 | 802 |
| QRQ55781.1 | 3.83e-164 | 3 | 814 | 4 | 802 |
| ALJ48972.1 | 3.83e-164 | 3 | 814 | 4 | 802 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8VYZ3 | 1.63e-09 | 457 | 683 | 84 | 292 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
| O23038 | 5.28e-09 | 468 | 682 | 100 | 298 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q9LVQ0 | 8.04e-09 | 562 | 683 | 93 | 210 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
| Q4PT34 | 2.70e-08 | 464 | 692 | 51 | 253 | Probable pectinesterase 56 OS=Arabidopsis thaliana OX=3702 GN=PME56 PE=2 SV=1 |
| Q9ZQA3 | 7.01e-08 | 468 | 660 | 100 | 265 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
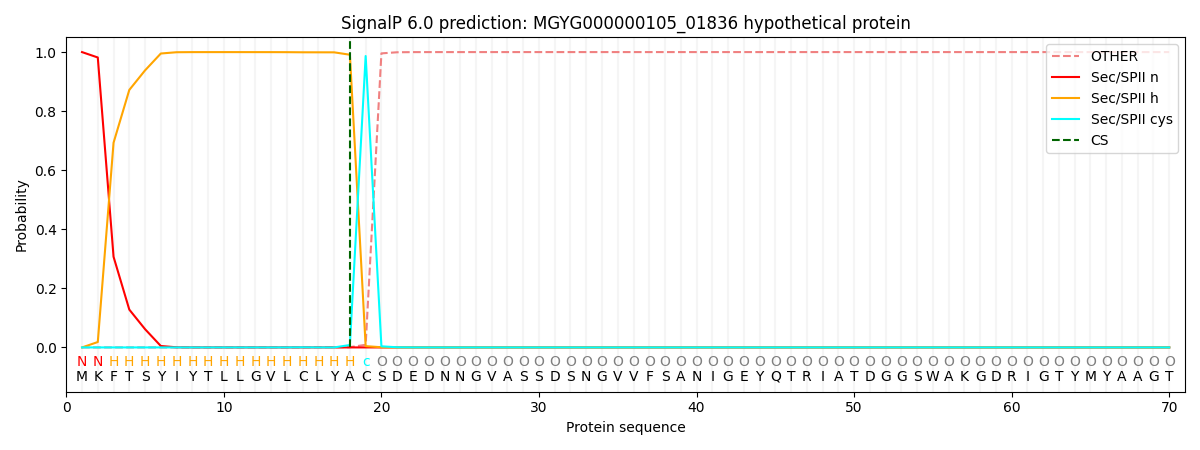
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000031 | 1.000037 | 0.000000 | 0.000000 | 0.000000 |
