You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000114_00131
You are here: Home > Sequence: MGYG000000114_00131
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
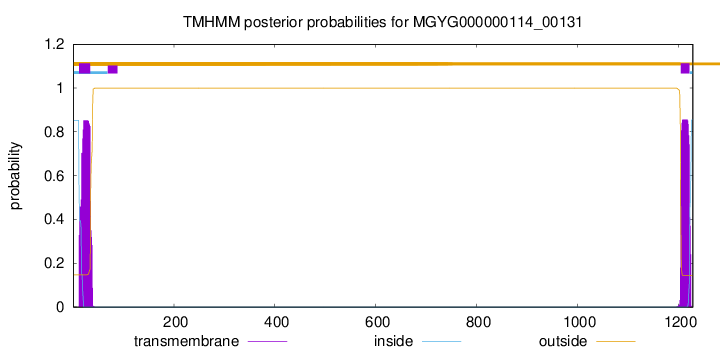
TMHMM annotations
Basic Information help
| Species | Streptococcus sp902363395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus sp902363395 | |||||||||||
| CAZyme ID | MGYG000000114_00131 | |||||||||||
| CAZy Family | GH29 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 150498; End: 154187 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH29 | 116 | 423 | 1.7e-69 | 0.861271676300578 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3669 | AfuC | 2.54e-103 | 112 | 555 | 8 | 430 | Alpha-L-fucosidase [Carbohydrate transport and metabolism]. |
| pfam01120 | Alpha_L_fucos | 6.13e-31 | 130 | 418 | 30 | 323 | Alpha-L-fucosidase. |
| smart00812 | Alpha_L_fucos | 1.43e-30 | 109 | 419 | 9 | 326 | Alpha-L-fucosidase. O-Glycosyl hydrolases (EC 3.2.1.-) are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycosyl hydrolases, based on sequence similarity, has led to the definition of 85 different families. This classification is available on the CAZy (CArbohydrate-Active EnZymes) web site. Because the fold of proteins is better conserved than their sequences, some of the families can be grouped in 'clans'. Family 29 encompasses alpha-L-fucosidases, which is a lysosomal enzyme responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. Deficiency of alpha-L-fucosidase results in the lysosomal storage disease fucosidosis. |
| pfam07501 | G5 | 1.03e-11 | 1058 | 1126 | 6 | 75 | G5 domain. This domain is found in a wide range of extracellular proteins. It is found tandemly repeated in up to 8 copies. It is found in the N-terminus of peptidases belonging to the M26 family which cleave human IgA. The domain is also found in proteins involved in metabolism of bacterial cell walls suggesting this domain may have an adhesive function. |
| TIGR01168 | YSIRK_signal | 3.84e-10 | 3 | 40 | 2 | 39 | Gram-positive signal peptide, YSIRK family. Many surface proteins found in Streptococcus, Staphylococcus, and related lineages share apparently homologous signal sequences. A motif resembling [YF]SIRKxxxGxxS[VIA] appears at the start of the transmembrane domain. The GxxS motif appears perfectly conserved, suggesting a specific function and not just homology. There is a strong correlation between proteins carrying this region at the N-terminus and those carrying the Gram-positive anchor domain with the LPXTG sortase processing site at the C-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGY39008.1 | 0.0 | 1 | 1229 | 1 | 1229 |
| AMP67381.1 | 0.0 | 1 | 1229 | 1 | 1229 |
| VEE18999.1 | 0.0 | 1 | 1229 | 1 | 1231 |
| VED68021.1 | 0.0 | 1 | 1229 | 1 | 1231 |
| SQH67270.1 | 0.0 | 1 | 1229 | 1 | 1231 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ORG_A | 2.19e-191 | 108 | 558 | 4 | 451 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORG_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 6OR4_A | 3.28e-190 | 108 | 558 | 4 | 451 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6OR4_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORH_A Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORH_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 6ORF_A | 3.40e-190 | 108 | 558 | 4 | 451 | Crystalstructure of SpGH29 [Streptococcus pneumoniae TIGR4],6ORF_B Crystal structure of SpGH29 [Streptococcus pneumoniae TIGR4] |
| 6TR3_A | 2.25e-112 | 114 | 602 | 27 | 544 | Ruminococcusgnavus GH29 fucosidase E1_10125 in complex with fucose [[Ruminococcus] gnavus E1] |
| 6TR4_A | 3.13e-111 | 114 | 602 | 27 | 544 | Ruminococcusgnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose [[Ruminococcus] gnavus E1],6TR4_B Ruminococcus gnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose [[Ruminococcus] gnavus E1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7XUR3 | 1.60e-106 | 113 | 561 | 38 | 482 | Putative alpha-L-fucosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0560400 PE=3 SV=2 |
| Q8GW72 | 1.57e-104 | 113 | 555 | 36 | 477 | Alpha-L-fucosidase 1 OS=Arabidopsis thaliana OX=3702 GN=FUC1 PE=1 SV=2 |
| P49610 | 2.65e-06 | 911 | 1213 | 1024 | 1298 | Beta-N-acetylhexosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=strH PE=1 SV=2 |
| P17164 | 3.81e-06 | 152 | 406 | 110 | 341 | Tissue alpha-L-fucosidase OS=Rattus norvegicus OX=10116 GN=Fuca1 PE=1 SV=1 |
| Q99LJ1 | 6.49e-06 | 161 | 406 | 104 | 331 | Tissue alpha-L-fucosidase OS=Mus musculus OX=10090 GN=Fuca1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
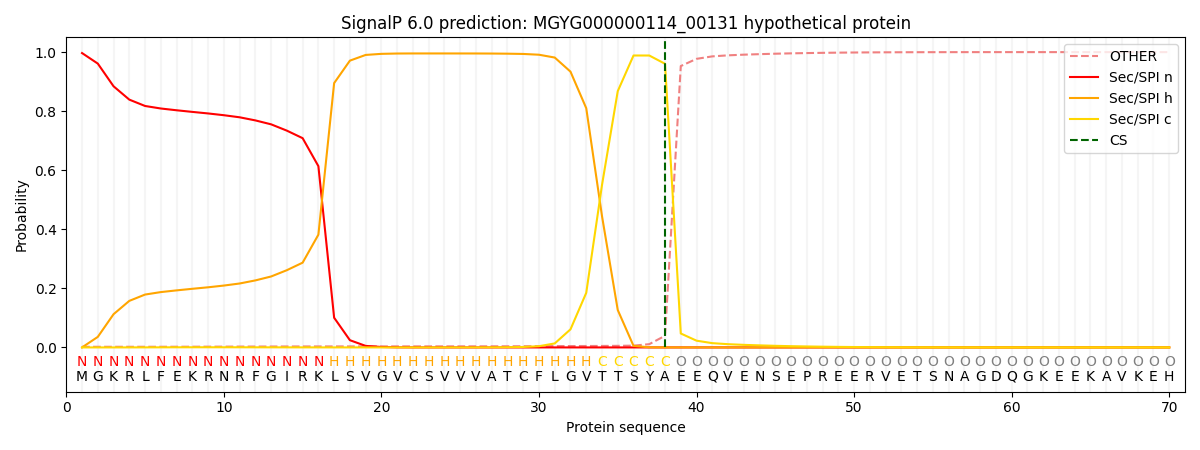
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003097 | 0.994630 | 0.001630 | 0.000241 | 0.000197 | 0.000178 |