You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000129_01506
You are here: Home > Sequence: MGYG000000129_01506
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
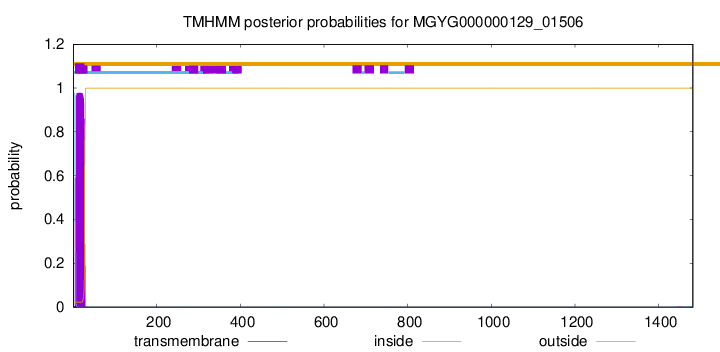
TMHMM annotations
Basic Information help
| Species | Marseille-P4683 sp900232885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Marseille-P4683; Marseille-P4683 sp900232885 | |||||||||||
| CAZyme ID | MGYG000000129_01506 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 60182; End: 64630 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 469 | 671 | 1.3e-52 | 0.9948453608247423 |
| CE1 | 822 | 1037 | 3.8e-32 | 0.9559471365638766 |
| CBM32 | 40 | 168 | 7.8e-25 | 0.9354838709677419 |
| CBM32 | 184 | 309 | 7.2e-23 | 0.9274193548387096 |
| CBM32 | 324 | 452 | 1.3e-21 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 2.27e-59 | 809 | 1042 | 158 | 386 | Predicted peptidase [General function prediction only]. |
| cd01833 | XynB_like | 5.79e-42 | 469 | 671 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 4.21e-22 | 474 | 660 | 2 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam00754 | F5_F8_type_C | 1.91e-16 | 183 | 309 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.37e-15 | 40 | 150 | 1 | 110 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61582.1 | 0.0 | 1 | 1482 | 1 | 1482 |
| BBH23072.1 | 8.78e-72 | 29 | 456 | 1089 | 1526 |
| QNK56923.1 | 1.06e-70 | 34 | 456 | 327 | 738 |
| BCI61304.1 | 1.98e-69 | 317 | 1481 | 1151 | 2271 |
| QHW32259.1 | 6.85e-68 | 32 | 467 | 89 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 5.03e-63 | 809 | 1041 | 142 | 378 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 3.32e-38 | 825 | 1037 | 21 | 211 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 2VPT_A | 2.06e-33 | 469 | 681 | 6 | 205 | ChainA, LIPOLYTIC ENZYME [Acetivibrio thermocellus] |
| 4Q82_A | 5.10e-20 | 830 | 1041 | 70 | 275 | CrystalStructure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365],4Q82_B Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum [Haliangium ochraceum DSM 14365] |
| 2RVA_A | 1.03e-08 | 1048 | 1184 | 7 | 136 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15329 | 7.87e-21 | 528 | 681 | 3 | 147 | Putative endoglucanase X (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celX PE=1 SV=1 |
| Q9RLB8 | 5.31e-18 | 469 | 680 | 44 | 269 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
| A1CC33 | 8.70e-07 | 827 | 1009 | 47 | 211 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| Q0CDX2 | 1.14e-06 | 812 | 1009 | 30 | 209 | Probable feruloyl esterase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
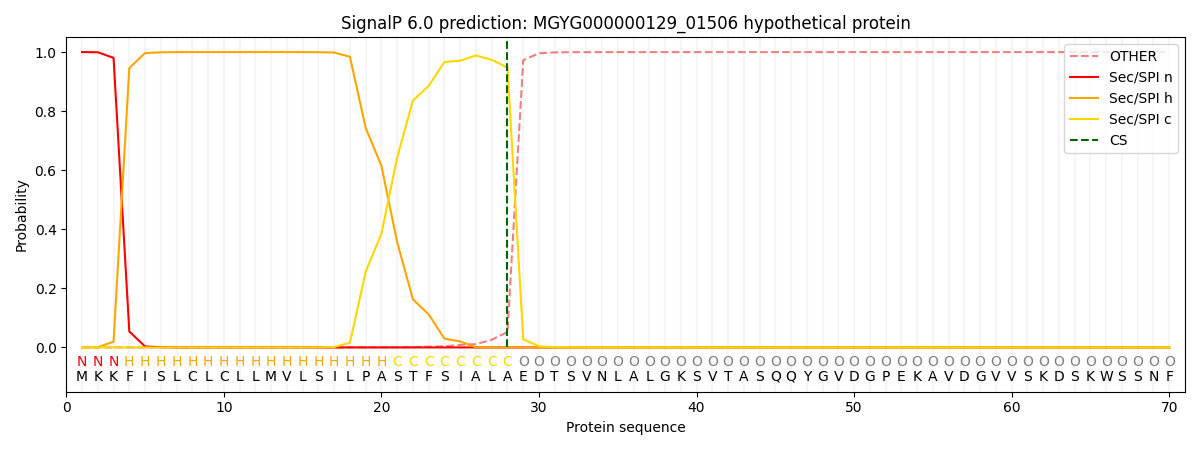
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000344 | 0.998878 | 0.000193 | 0.000206 | 0.000179 | 0.000167 |