You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000135_03018
Basic Information
help
| Species |
Murimonas intestini
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Murimonas; Murimonas intestini
|
| CAZyme ID |
MGYG000000135_03018
|
| CAZy Family |
GH142 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000135 |
5745007 |
Isolate |
Canada |
North America |
|
| Gene Location |
Start: 205434;
End: 206912
Strand: +
|
No EC number prediction in MGYG000000135_03018.
| Family |
Start |
End |
Evalue |
family coverage |
| GH142 |
14 |
488 |
7e-153 |
0.9686847599164927 |
MGYG000000135_03018 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5MQS_A
|
1.48e-101 |
10 |
488 |
628 |
1086 |
SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQR_A
|
3.99e-97 |
10 |
488 |
628 |
1086 |
SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A
|
1.76e-12 |
35 |
487 |
305 |
793 |
Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
E8MGH9
|
1.20e-11 |
35 |
487 |
336 |
824 |
Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
This protein is predicted as OTHER
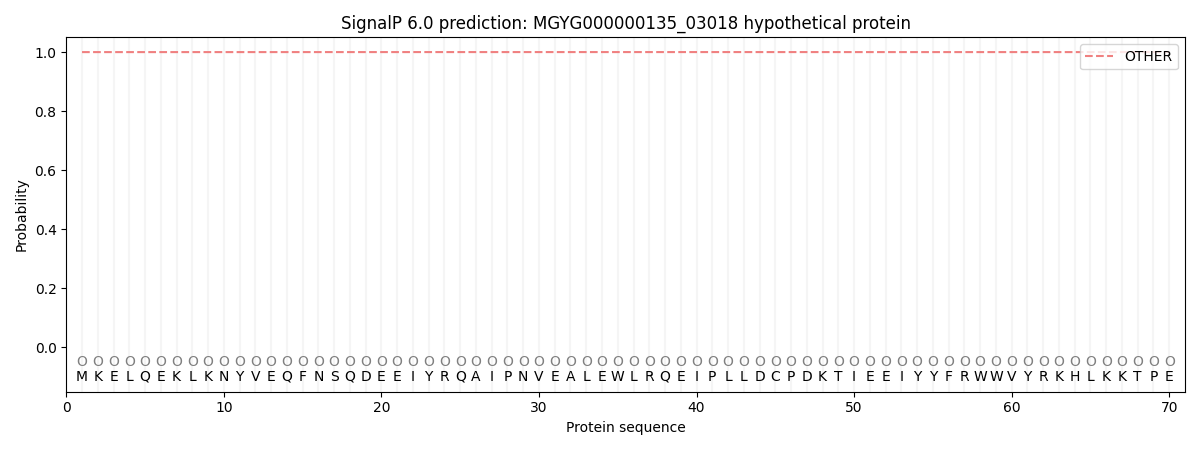
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000044
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000000135_03018.

