You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000149_00562
You are here: Home > Sequence: MGYG000000149_00562
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
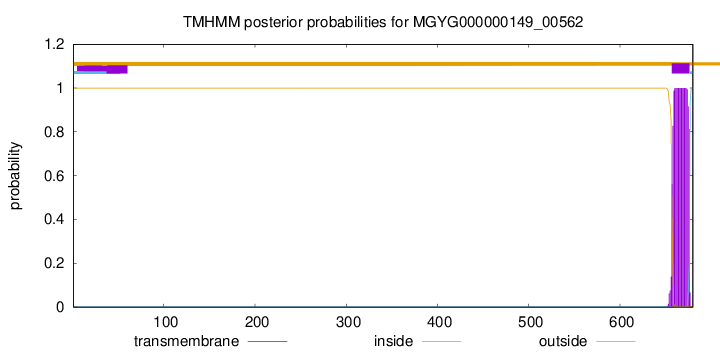
TMHMM annotations
Basic Information help
| Species | Clostridium saudiense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium saudiense | |||||||||||
| CAZyme ID | MGYG000000149_00562 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 17151; End: 19193 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 104 | 401 | 3.6e-86 | 0.9618055555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 3.85e-52 | 104 | 402 | 90 | 402 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 1.35e-51 | 107 | 397 | 11 | 290 | Pectinesterase. |
| PLN02432 | PLN02432 | 1.00e-46 | 105 | 402 | 20 | 285 | putative pectinesterase |
| PLN02682 | PLN02682 | 3.82e-45 | 86 | 380 | 69 | 336 | pectinesterase family protein |
| PLN02671 | PLN02671 | 2.38e-39 | 47 | 396 | 18 | 345 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CED92996.1 | 1.74e-252 | 1 | 600 | 602 | 1206 |
| QJS18268.1 | 7.21e-170 | 1 | 434 | 437 | 875 |
| BCS80168.1 | 7.26e-89 | 4 | 402 | 842 | 1241 |
| ADQ45070.1 | 7.31e-85 | 4 | 397 | 842 | 1236 |
| ADQ05621.1 | 6.95e-81 | 4 | 397 | 843 | 1237 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 3.89e-36 | 108 | 402 | 44 | 358 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 2NSP_A | 2.15e-35 | 94 | 409 | 4 | 341 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 1.21e-31 | 94 | 409 | 4 | 341 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 1.21e-31 | 94 | 409 | 4 | 341 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 5C1E_A | 4.92e-28 | 91 | 397 | 1 | 284 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A9 | 1.05e-30 | 94 | 409 | 28 | 365 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| P0C1A8 | 1.43e-30 | 94 | 409 | 28 | 365 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| Q9FM79 | 1.73e-28 | 110 | 380 | 94 | 347 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| O23038 | 1.29e-27 | 108 | 382 | 101 | 359 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q8L7Q7 | 1.36e-27 | 108 | 379 | 301 | 566 | Probable pectinesterase/pectinesterase inhibitor 64 OS=Arabidopsis thaliana OX=3702 GN=PME64 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
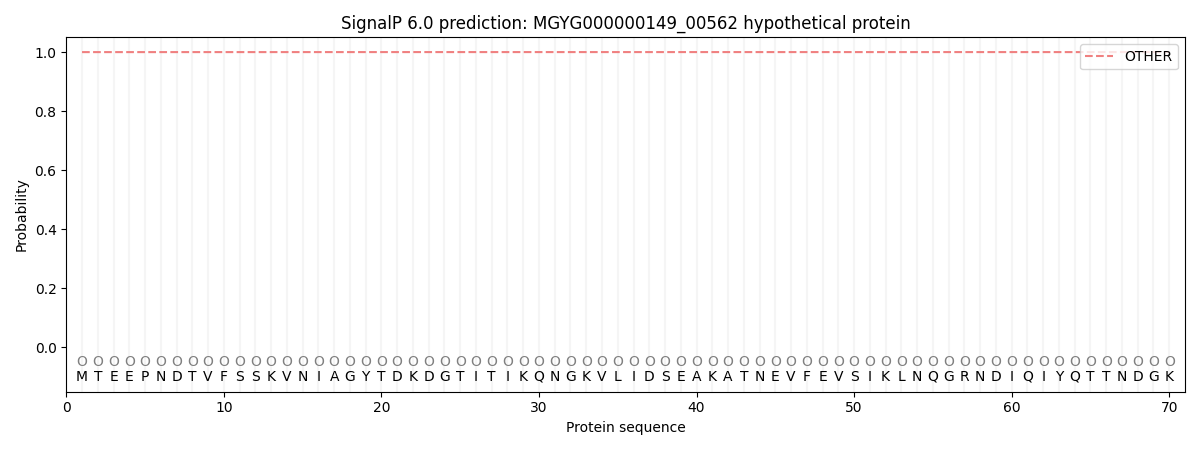
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000041 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |