You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000169_01699
You are here: Home > Sequence: MGYG000000169_01699
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp000432355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp000432355 | |||||||||||
| CAZyme ID | MGYG000000169_01699 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 148721; End: 153940 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 63 | 460 | 2.2e-68 | 0.9695945945945946 |
| GH18 | 1192 | 1514 | 3.7e-20 | 0.9662162162162162 |
| GH18 | 747 | 1038 | 1.5e-19 | 0.9628378378378378 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 3.99e-105 | 66 | 451 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd02871 | GH18_chitinase_D-like | 1.92e-100 | 746 | 1042 | 1 | 311 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| COG3325 | ChiA | 5.67e-94 | 64 | 464 | 38 | 436 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
| cd02871 | GH18_chitinase_D-like | 4.10e-91 | 1193 | 1517 | 4 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| smart00636 | Glyco_18 | 4.76e-86 | 65 | 451 | 1 | 334 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS39875.1 | 2.75e-275 | 60 | 1521 | 39 | 1373 |
| QXF14593.1 | 3.95e-194 | 68 | 619 | 1 | 543 |
| QXF14594.1 | 3.18e-142 | 132 | 619 | 7 | 470 |
| AQM59350.1 | 3.55e-115 | 58 | 751 | 45 | 765 |
| AIY83966.1 | 8.07e-114 | 58 | 751 | 45 | 765 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EBV_A | 3.97e-78 | 745 | 1044 | 4 | 291 | Crystalstructure of putative Chitinase A from Streptomyces coelicolor. [Streptomyces coelicolor] |
| 3N11_A | 2.42e-70 | 1187 | 1506 | 4 | 315 | Crystalstricture of wild-type chitinase from Bacillus cereus NCTU2 [Bacillus cereus],3N12_A Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N15_A | 4.48e-70 | 1187 | 1506 | 4 | 315 | Crystalstricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
| 5KZ6_A | 6.67e-70 | 1187 | 1506 | 7 | 318 | 1.25Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis],5KZ6_B 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis] |
| 3N17_A | 1.53e-69 | 1187 | 1506 | 4 | 315 | Crystalstricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 2.38e-70 | 1084 | 1521 | 98 | 514 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| A5FB63 | 3.25e-68 | 67 | 1509 | 28 | 1469 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| Q05638 | 1.79e-61 | 1175 | 1502 | 249 | 581 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
| D4AVJ0 | 2.13e-58 | 1209 | 1521 | 17 | 331 | Probable class II chitinase ARB_00204 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00204 PE=1 SV=2 |
| P20533 | 5.69e-55 | 53 | 711 | 33 | 697 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
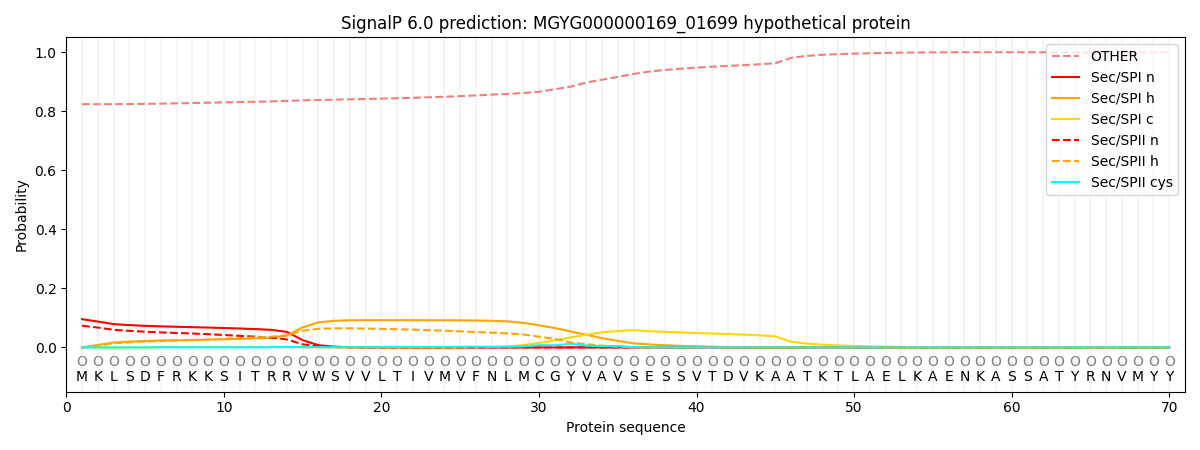
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.827453 | 0.089810 | 0.074479 | 0.000374 | 0.000217 | 0.007667 |

