You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_02050
You are here: Home > Sequence: MGYG000000174_02050
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_02050 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 364300; End: 366117 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 34 | 517 | 1.8e-72 | 0.8222222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 4.87e-27 | 35 | 412 | 13 | 369 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 1.03e-04 | 86 | 247 | 2 | 160 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAJ74247.1 | 1.76e-30 | 48 | 406 | 28 | 369 |
| AFY78130.1 | 2.10e-24 | 47 | 394 | 27 | 347 |
| AXL21031.1 | 7.10e-23 | 24 | 401 | 3 | 349 |
| AGA77175.1 | 1.02e-21 | 57 | 379 | 37 | 336 |
| QSV72899.1 | 1.59e-21 | 43 | 401 | 16 | 350 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 3.18e-07 | 57 | 374 | 59 | 358 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A8FRR0 | 8.70e-13 | 47 | 410 | 24 | 368 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
| O67270 | 1.36e-12 | 28 | 379 | 1 | 320 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| O67601 | 2.33e-12 | 51 | 401 | 24 | 325 | Uncharacterized protein aq_1704 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1704 PE=3 SV=1 |
| B2VBI7 | 2.35e-10 | 57 | 392 | 32 | 344 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| A4WAM1 | 5.01e-09 | 57 | 380 | 32 | 332 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Enterobacter sp. (strain 638) OX=399742 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999755 | 0.000258 | 0.000002 | 0.000000 | 0.000000 | 0.000005 |
TMHMM Annotations download full data without filtering help
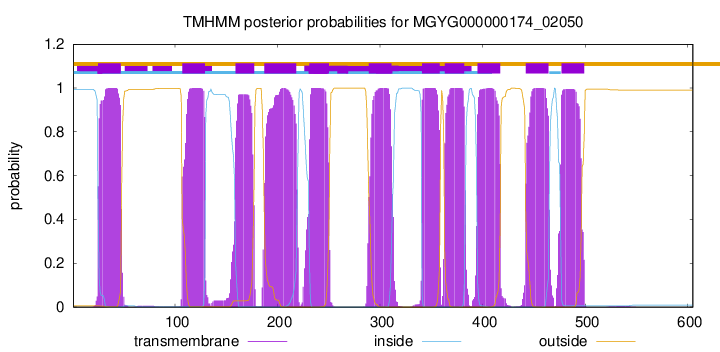
| start | end |
|---|---|
| 25 | 47 |
| 107 | 129 |
| 159 | 177 |
| 187 | 218 |
| 231 | 250 |
| 289 | 311 |
| 341 | 358 |
| 363 | 382 |
| 395 | 417 |
| 442 | 464 |
| 477 | 499 |
