You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000174_02518
You are here: Home > Sequence: MGYG000000174_02518
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
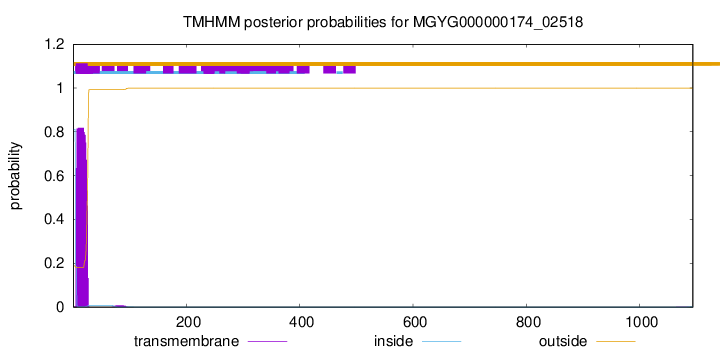
TMHMM annotations
Basic Information help
| Species | Parabacteroides faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides faecis | |||||||||||
| CAZyme ID | MGYG000000174_02518 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 194168; End: 197452 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 342 | 1087 | 2.5e-210 | 0.8847087378640777 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 37 | 885 | 1 | 866 | alpha-L-rhamnosidase. |
| pfam00754 | F5_F8_type_C | 1.05e-10 | 181 | 280 | 1 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 5.56e-05 | 180 | 277 | 12 | 127 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 6.23e-05 | 183 | 277 | 16 | 122 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| cd08667 | APC10-ZZEF1 | 8.96e-04 | 182 | 219 | 7 | 44 | APC10/DOC1-like domain of uncharacterized Zinc finger ZZ-type and EF-hand domain-containing protein 1 (ZZEF1) and homologs. This model represents the APC10/DOC1-like domain present in the uncharacterized Zinc finger ZZ-type and EF-hand domain-containing protein 1 (ZZEF1) of Mus musculus. Members of this family contain EF-hand, APC10, CUB, and zinc finger ZZ-type domains. ZZEF1-like APC10 domains are homologous to the APC10 subunit/DOC1 domains present in E3 ubiquitin ligases, which mediate substrate ubiquitination (or ubiquitylation), and are components of the ubiquitin-26S proteasome pathway for selective proteolytic degradation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV99811.1 | 1.41e-245 | 34 | 1088 | 42 | 1006 |
| ACU05683.1 | 1.13e-241 | 36 | 1087 | 50 | 985 |
| QEH40462.1 | 2.81e-241 | 35 | 1090 | 44 | 992 |
| AWO01978.1 | 2.40e-239 | 20 | 1085 | 31 | 999 |
| AWI09582.1 | 7.20e-236 | 34 | 1086 | 44 | 1025 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 4.18e-128 | 36 | 1089 | 46 | 1141 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 3.33e-105 | 34 | 1093 | 33 | 1105 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 8.70e-105 | 34 | 1093 | 33 | 1105 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6D8K_A | 1.64e-06 | 967 | 1048 | 88 | 183 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 8.41e-84 | 31 | 1057 | 28 | 1102 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000333 | 0.998990 | 0.000174 | 0.000175 | 0.000154 | 0.000145 |