You are browsing environment: HUMAN GUT
MGYG000000182_00595
Basic Information
help
Species
Mitsuokella sp003458855
Lineage
Bacteria; Firmicutes_C; Negativicutes; Selenomonadales; Selenomonadaceae; Mitsuokella; Mitsuokella sp003458855
CAZyme ID
MGYG000000182_00595
CAZy Family
GH73
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000182
2580428
Isolate
China
Asia
Gene Location
Start: 64403;
End: 65242
Strand: -
No EC number prediction in MGYG000000182_00595.
Family
Start
End
Evalue
family coverage
GH73
149
274
7.7e-16
0.9453125
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam01832
Glucosaminidase
5.86e-12
150
214
3
77
Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O51481
5.72e-21
112
259
37
179
Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1
more
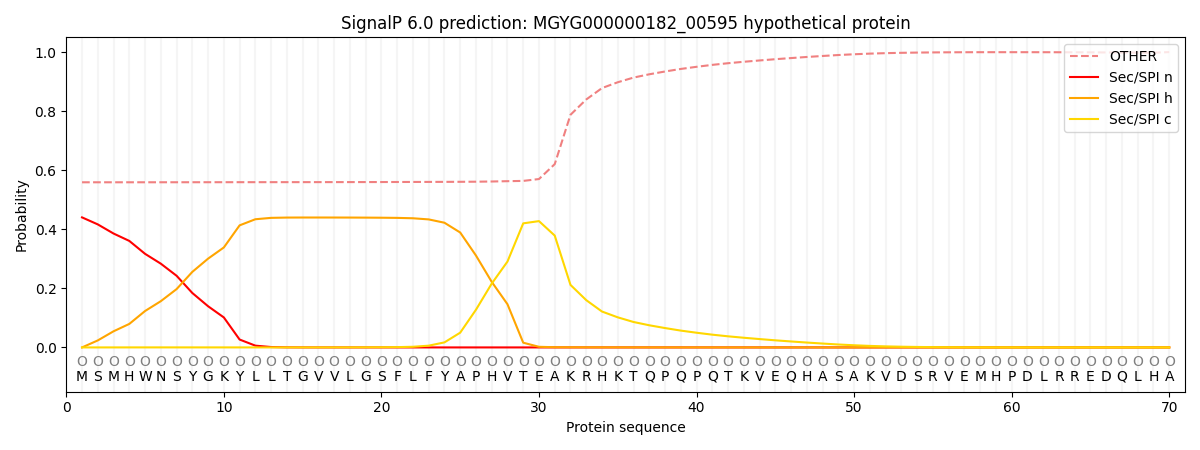
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.578399
0.419437
0.001090
0.000312
0.000292
0.000475