You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000201_00358
You are here: Home > Sequence: MGYG000000201_00358
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
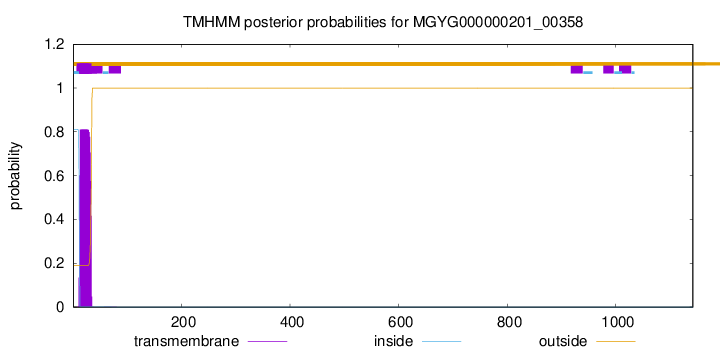
TMHMM annotations
Basic Information help
| Species | Blautia_A sp900066145 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp900066145 | |||||||||||
| CAZyme ID | MGYG000000201_00358 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 414730; End: 418158 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 119 | 314 | 5.3e-26 | 0.7962962962962963 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 6.23e-38 | 116 | 973 | 111 | 758 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 1.40e-28 | 690 | 862 | 85 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PLN03080 | PLN03080 | 2.29e-23 | 122 | 939 | 111 | 742 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 5.98e-22 | 117 | 471 | 75 | 365 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam14310 | Fn3-like | 1.76e-19 | 897 | 969 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYA77556.1 | 1.32e-275 | 39 | 1136 | 35 | 1140 |
| QKS56187.1 | 4.59e-259 | 41 | 1139 | 35 | 1142 |
| QMV43973.1 | 1.79e-245 | 40 | 1136 | 9 | 1111 |
| AYQ75660.1 | 1.79e-231 | 18 | 1142 | 14 | 1135 |
| QOS79412.1 | 1.99e-182 | 58 | 1140 | 31 | 1122 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AC0_A | 3.92e-32 | 106 | 980 | 48 | 836 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 3ABZ_A | 5.74e-30 | 106 | 980 | 48 | 836 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
| 3ZYZ_A | 2.84e-26 | 654 | 980 | 385 | 712 | ChainA, Beta-d-glucoside Glucohydrolase [Trichoderma reesei],3ZZ1_A Chain A, BETA-D-GLUCOSIDE GLUCOHYDROLASE [Trichoderma reesei] |
| 4I8D_A | 2.84e-26 | 654 | 980 | 386 | 713 | ChainA, Beta-D-glucoside glucohydrolase [Trichoderma reesei],4I8D_B Chain B, Beta-D-glucoside glucohydrolase [Trichoderma reesei] |
| 7MS2_A | 2.24e-25 | 696 | 972 | 404 | 660 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 1.88e-29 | 113 | 971 | 73 | 851 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| P27034 | 2.04e-28 | 115 | 980 | 50 | 810 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| Q5B0F4 | 5.66e-27 | 711 | 973 | 540 | 809 | Probable beta-glucosidase G OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglG PE=3 SV=2 |
| Q0CAF5 | 5.91e-27 | 125 | 978 | 64 | 829 | Probable beta-glucosidase I OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bglI PE=3 SV=1 |
| Q2UNR0 | 8.25e-27 | 651 | 973 | 430 | 741 | Probable beta-glucosidase D OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
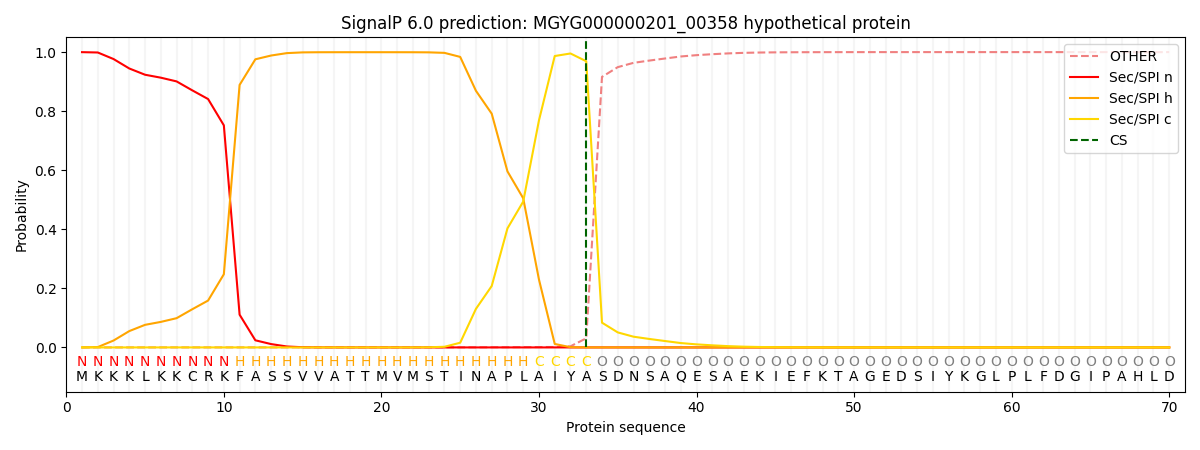
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000580 | 0.998508 | 0.000278 | 0.000263 | 0.000188 | 0.000153 |