You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000201_01942
You are here: Home > Sequence: MGYG000000201_01942
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Blautia_A sp900066145 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; Blautia_A sp900066145 | |||||||||||
| CAZyme ID | MGYG000000201_01942 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 246474; End: 248507 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 5 | 126 | 6.7e-35 | 0.7235294117647059 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03025 | EPS_sugtrans | 2.76e-51 | 249 | 675 | 8 | 442 | exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase. Members of this family are generally found near other genes involved in the biosynthesis of a variety of exopolysaccharides. These proteins consist of two fused domains, an N-terminal hydrophobic domain of generally low conservation and a highly conserved C-terminal sugar transferase domain (pfam02397). Characterized and partially characterized members of this subfamily include Salmonella WbaP (originally RfbP), E. coli WcaJ, Methylobacillus EpsB, Xanthomonas GumD, Vibrio CpsA, Erwinia AmsG, Group B Streptococcus CpsE (originally CpsD), and Streptococcus suis Cps2E. Each of these is believed to act in transferring the sugar from, for instance, UDP-glucose or UDP-galactose, to a lipid carrier such as undecaprenyl phosphate as the first (priming) step in the synthesis of an oligosaccharide "block". This function is encoded in the C-terminal domain. The liposaccharide is believed to be subsequently transferred through a "flippase" function from the cytoplasmic to the periplasmic face of the inner membrane by the N-terminal domain. Certain closely related transferase enzymes, such as Sinorhizobium ExoY and Lactococcus EpsD, lack the N-terminal domain and are not found by this model. |
| pfam02397 | Bac_transf | 2.28e-50 | 516 | 672 | 1 | 180 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| COG2148 | WcaJ | 1.70e-39 | 508 | 672 | 33 | 220 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| pfam00535 | Glycos_transf_2 | 5.12e-37 | 5 | 162 | 1 | 163 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
| cd00761 | Glyco_tranf_GTA_type | 7.66e-35 | 6 | 123 | 1 | 118 | Glycosyltransferase family A (GT-A) includes diverse families of glycosyl transferases with a common GT-A type structural fold. Glycosyltransferases (GTs) are enzymes that synthesize oligosaccharides, polysaccharides, and glycoconjugates by transferring the sugar moiety from an activated nucleotide-sugar donor to an acceptor molecule, which may be a growing oligosaccharide, a lipid, or a protein. Based on the stereochemistry of the donor and acceptor molecules, GTs are classified as either retaining or inverting enzymes. To date, all GT structures adopt one of two possible folds, termed GT-A fold and GT-B fold. This hierarchy includes diverse families of glycosyl transferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. The majority of the proteins in this superfamily are Glycosyltransferase family 2 (GT-2) proteins. But it also includes families GT-43, GT-6, GT-8, GT13 and GT-7; which are evolutionarily related to GT-2 and share structure similarities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL24103.1 | 0.0 | 1 | 674 | 1 | 690 |
| QUL53905.1 | 4.32e-72 | 4 | 249 | 5 | 248 |
| AUM96479.1 | 4.09e-71 | 2 | 248 | 9 | 253 |
| AVQ53931.1 | 4.09e-71 | 2 | 248 | 9 | 253 |
| QNO13385.1 | 5.50e-70 | 4 | 249 | 8 | 251 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BCV_A | 2.50e-17 | 1 | 244 | 4 | 223 | Crystalstructure of a putative glycosyltransferase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3BCV_B Crystal structure of a putative glycosyltransferase from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 2Z87_A | 4.03e-17 | 1 | 189 | 373 | 559 | Crystalstructure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP [Escherichia coli],2Z87_B Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP [Escherichia coli] |
| 2Z86_A | 4.04e-17 | 1 | 189 | 374 | 560 | Crystalstructure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_B Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_C Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli],2Z86_D Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP [Escherichia coli] |
| 5W7L_A | 3.53e-16 | 507 | 669 | 2 | 182 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
| 5HEA_A | 3.83e-16 | 4 | 125 | 7 | 127 | CgTstructure in hexamer [Streptococcus parasanguinis FW213],5HEA_B CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEA_C CgT structure in hexamer [Streptococcus parasanguinis FW213],5HEC_A CgT structure in dimer [Streptococcus parasanguinis FW213],5HEC_B CgT structure in dimer [Streptococcus parasanguinis FW213] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O32268 | 7.46e-46 | 2 | 223 | 6 | 225 | Putative teichuronic acid biosynthesis glycosyltransferase TuaG OS=Bacillus subtilis (strain 168) OX=224308 GN=tuaG PE=2 SV=1 |
| Q077R2 | 3.54e-30 | 1 | 248 | 1 | 241 | UDP-Glc:alpha-D-GlcNAc-diphosphoundecaprenol beta-1,3-glucosyltransferase WfaP OS=Escherichia coli OX=562 GN=wfaP PE=1 SV=1 |
| Q57022 | 4.71e-30 | 1 | 249 | 3 | 249 | Uncharacterized glycosyltransferase HI_0868 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0868 PE=3 SV=1 |
| B5L3F2 | 1.28e-29 | 4 | 249 | 6 | 245 | UDP-Glc:alpha-D-GlcNAc-diphosphoundecaprenol beta-1,3-glucosyltransferase WfgD OS=Escherichia coli OX=562 GN=wfgD PE=1 SV=1 |
| P71054 | 1.68e-25 | 2 | 203 | 5 | 208 | Putative glycosyltransferase EpsE OS=Bacillus subtilis (strain 168) OX=224308 GN=epsE PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
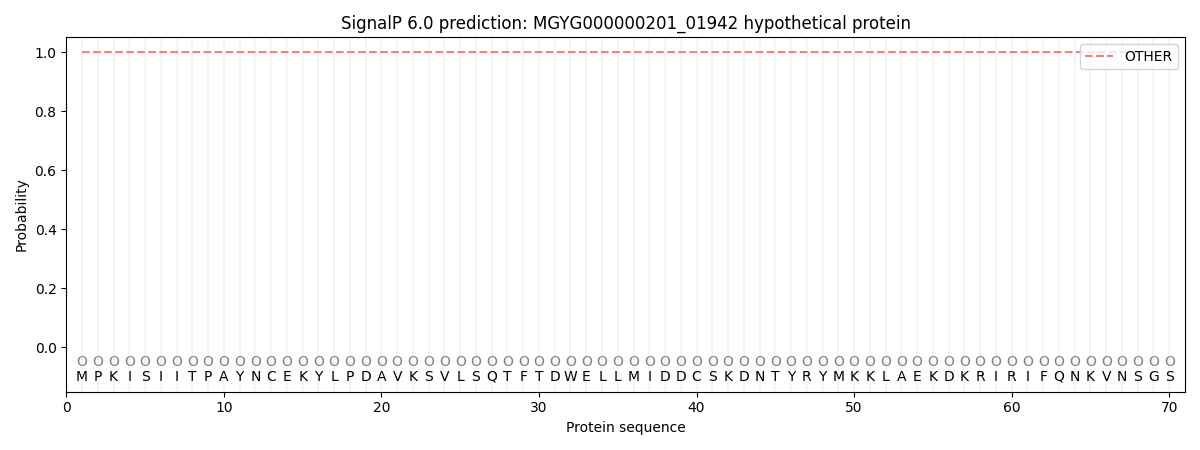
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000063 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
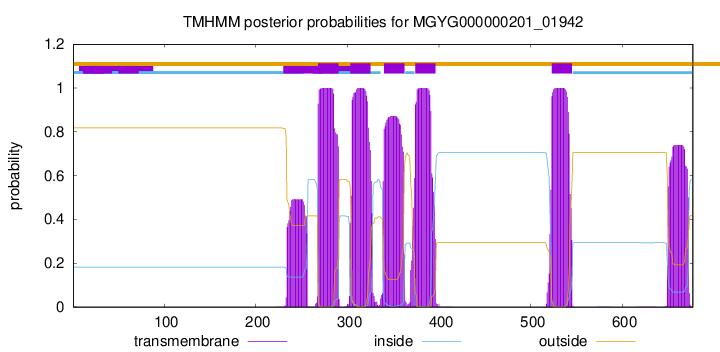
| start | end |
|---|---|
| 268 | 290 |
| 303 | 325 |
| 340 | 362 |
| 374 | 396 |
| 523 | 545 |
