You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000265_01040
You are here: Home > Sequence: MGYG000000265_01040
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides nordii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides nordii | |||||||||||
| CAZyme ID | MGYG000000265_01040 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 158340; End: 161381 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 634 | 948 | 1.6e-92 | 0.9603960396039604 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.11e-102 | 627 | 947 | 1 | 308 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.37e-93 | 672 | 945 | 1 | 261 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 6.99e-76 | 640 | 945 | 39 | 335 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADX05746.1 | 2.99e-103 | 684 | 985 | 7 | 308 |
| ADX05712.1 | 1.78e-74 | 618 | 926 | 60 | 380 |
| AUG58840.1 | 1.70e-66 | 625 | 953 | 275 | 597 |
| QBI56661.1 | 7.84e-66 | 625 | 951 | 47 | 359 |
| ACP87340.1 | 3.26e-65 | 631 | 958 | 4 | 315 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XYZ_A | 1.44e-66 | 627 | 944 | 28 | 337 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 6FHE_A | 2.34e-63 | 638 | 947 | 23 | 338 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 3W24_A | 1.48e-59 | 638 | 960 | 19 | 335 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W25_A | 9.73e-59 | 638 | 960 | 19 | 335 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W26_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 3W27_A | 9.73e-59 | 638 | 960 | 19 | 335 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W28_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3W29_A The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 2.00e-61 | 627 | 944 | 518 | 827 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q00177 | 1.30e-56 | 651 | 944 | 51 | 319 | Endo-1,4-beta-xylanase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnC PE=1 SV=1 |
| P23360 | 8.98e-56 | 652 | 953 | 55 | 329 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| Q4P902 | 1.86e-55 | 630 | 951 | 33 | 339 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| P38535 | 2.25e-54 | 638 | 982 | 220 | 563 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
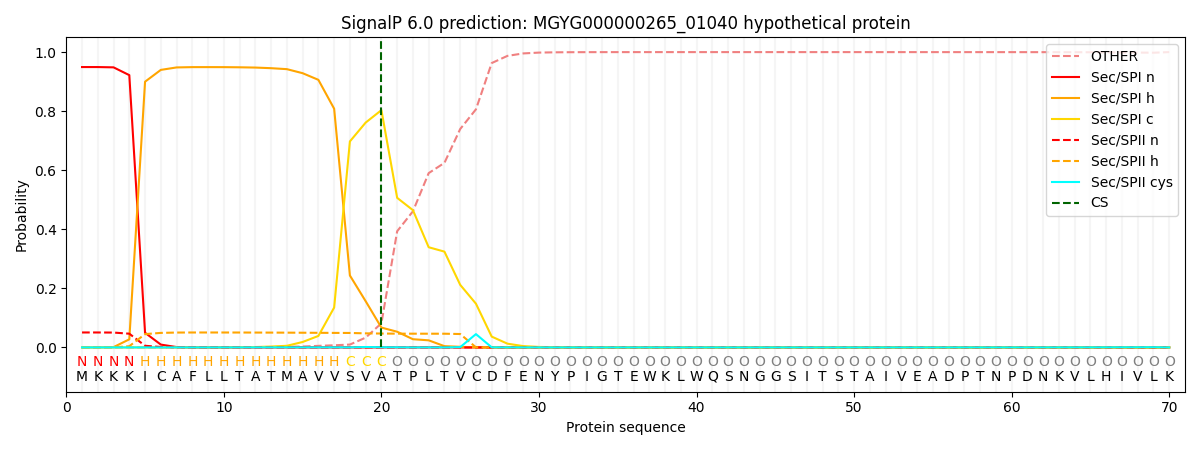
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001907 | 0.942564 | 0.054557 | 0.000348 | 0.000338 | 0.000288 |
