You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000273_00029
You are here: Home > Sequence: MGYG000000273_00029
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
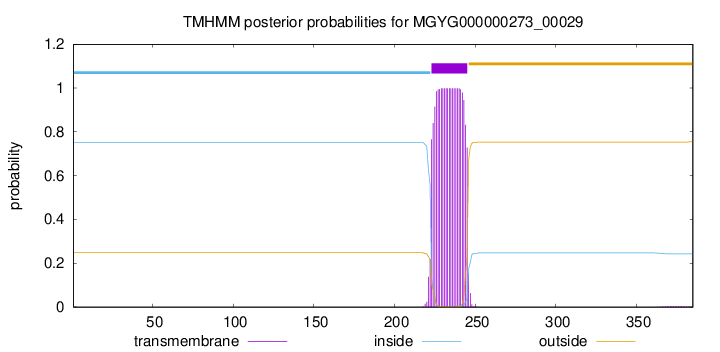
TMHMM annotations
Basic Information help
| Species | Phocaeicola coprophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola coprophilus | |||||||||||
| CAZyme ID | MGYG000000273_00029 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26768; End: 27925 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00216 | Bac_DNA_binding | 4.97e-08 | 10 | 77 | 5 | 88 | Bacterial DNA-binding protein. |
| cd00118 | LysM | 3.11e-07 | 328 | 377 | 1 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| smart00257 | LysM | 1.69e-06 | 329 | 377 | 1 | 44 | Lysin motif. |
| PRK11198 | PRK11198 | 2.08e-06 | 271 | 378 | 26 | 146 | LysM domain/BON superfamily protein; Provisional |
| cd00591 | HU_IHF | 2.40e-06 | 10 | 75 | 4 | 85 | DNA sequence specific (IHF) and non-specific (HU) domains. This family includes integration host factor (IHF) and HU, also called type II DNA-binding proteins (DNABII), which are small dimeric proteins that specifically bind the DNA minor groove, inducing large bends in the DNA and serving as architectural factors in a variety of cellular processes such as recombination, initiation of replication/transcription and gene regulation. IHF binds DNA in a sequence specific manner while HU displays little or no sequence preference. IHF homologs are usually heterodimers, while HU homologs are typically homodimers (except HU heterodimers from E. coli and other enterobacteria). HU is highly basic and contributes to chromosomal compaction and maintenance of negative supercoiling, thus often referred to as histone-like protein. IHF is an essential cofactor in phage lambda site-specific recombination, having an architectural role during assembly of specialized nucleoprotein structures (snups). Bacillus phage SPO1-encoded transcription factor 1 (TF1) is another related type II DNA-binding protein. Like IHF, TF1 binds DNA specifically and bends DNA sharply. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO25821.1 | 6.49e-251 | 1 | 385 | 1 | 385 |
| QQY44100.1 | 7.44e-57 | 1 | 382 | 1 | 474 |
| QQR17553.1 | 2.46e-52 | 1 | 380 | 1 | 441 |
| ANU57570.1 | 2.46e-52 | 1 | 380 | 1 | 441 |
| QCQ46347.1 | 2.15e-51 | 1 | 381 | 1 | 465 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
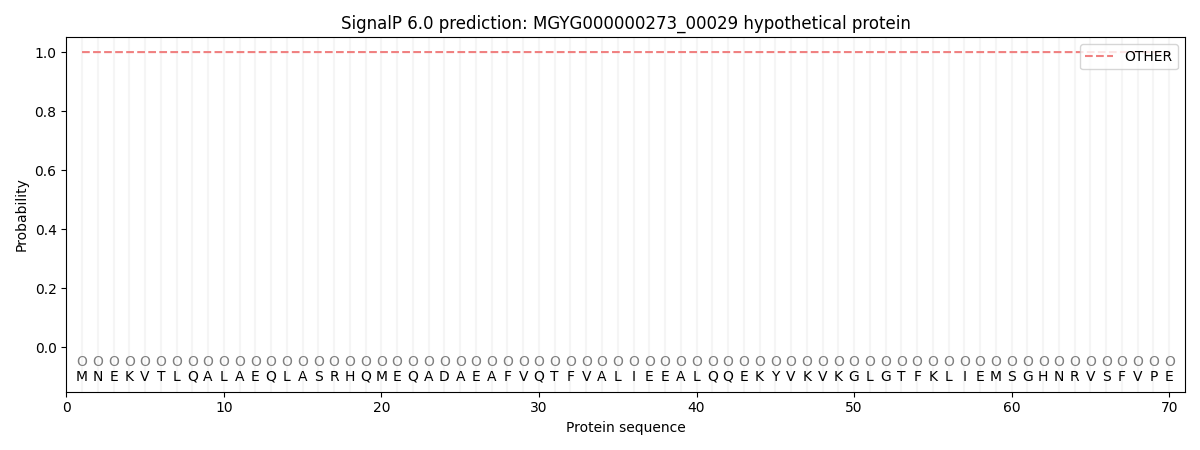
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000069 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |