You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000289_01139
You are here: Home > Sequence: MGYG000000289_01139
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella buccae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella buccae | |||||||||||
| CAZyme ID | MGYG000000289_01139 | |||||||||||
| CAZy Family | GH143 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 59112; End: 62564 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH143 | 51 | 595 | 5.2e-256 | 0.9892086330935251 |
| GH142 | 658 | 1143 | 1e-203 | 0.9937369519832986 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13088 | BNR_2 | 6.70e-09 | 294 | 368 | 168 | 247 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam01204 | Trehalase | 4.50e-05 | 868 | 917 | 308 | 357 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| cd15482 | Sialidase_non-viral | 0.007 | 296 | 368 | 207 | 289 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85039.1 | 0.0 | 58 | 1149 | 25 | 1111 |
| QUT58490.1 | 0.0 | 50 | 1149 | 19 | 1100 |
| QEW37190.1 | 0.0 | 50 | 1149 | 19 | 1100 |
| QQY37484.1 | 0.0 | 50 | 1149 | 19 | 1100 |
| ABR40380.1 | 0.0 | 50 | 1149 | 19 | 1100 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQR_A | 0.0 | 53 | 1149 | 25 | 1104 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQS_A | 0.0 | 53 | 1149 | 25 | 1104 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 6M5A_A | 3.38e-24 | 678 | 1136 | 304 | 799 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 3.12e-23 | 678 | 1136 | 335 | 830 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
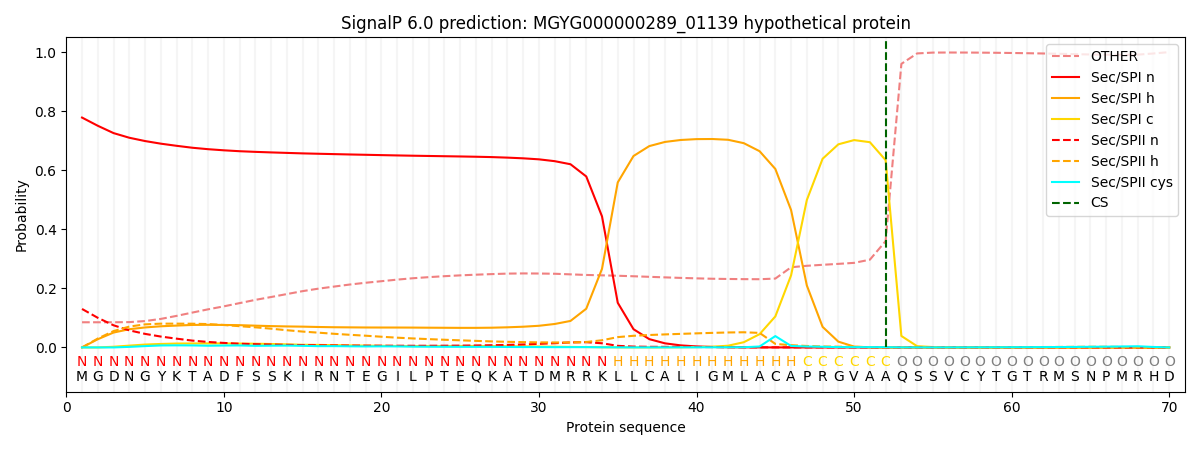
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.090989 | 0.734653 | 0.164510 | 0.006363 | 0.002765 | 0.000713 |
