You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000360_00954
You are here: Home > Sequence: MGYG000000360_00954
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900548385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900548385 | |||||||||||
| CAZyme ID | MGYG000000360_00954 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 231838; End: 234999 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 364 | 570 | 6.8e-24 | 0.45243619489559167 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 6.27e-10 | 356 | 511 | 69 | 218 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 1.09e-07 | 366 | 523 | 74 | 226 | Glycosyl hydrolases family 39. |
| pfam00331 | Glyco_hydro_10 | 1.16e-07 | 356 | 485 | 46 | 164 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.14e-05 | 356 | 485 | 3 | 124 | Glycosyl hydrolase family 10. |
| pfam04234 | CopC | 0.002 | 244 | 300 | 26 | 79 | CopC domain. CopC is a bacterial blue copper protein that binds 1 atom of copper per protein molecule. Along with CopA, CopC mediates copper resistance by sequestration of copper in the periplasm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44457.1 | 7.17e-299 | 16 | 1050 | 17 | 1063 |
| AVM45381.1 | 1.78e-150 | 9 | 1041 | 10 | 1036 |
| AHF89348.1 | 4.35e-21 | 342 | 553 | 405 | 628 |
| QNJ91947.1 | 4.72e-18 | 356 | 555 | 1123 | 1309 |
| BBH21833.1 | 5.57e-18 | 356 | 553 | 39 | 231 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WUF_A | 3.46e-07 | 319 | 517 | 4 | 197 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUB_A | 8.14e-07 | 319 | 517 | 4 | 197 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4XVN1 | 7.26e-06 | 319 | 517 | 42 | 235 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
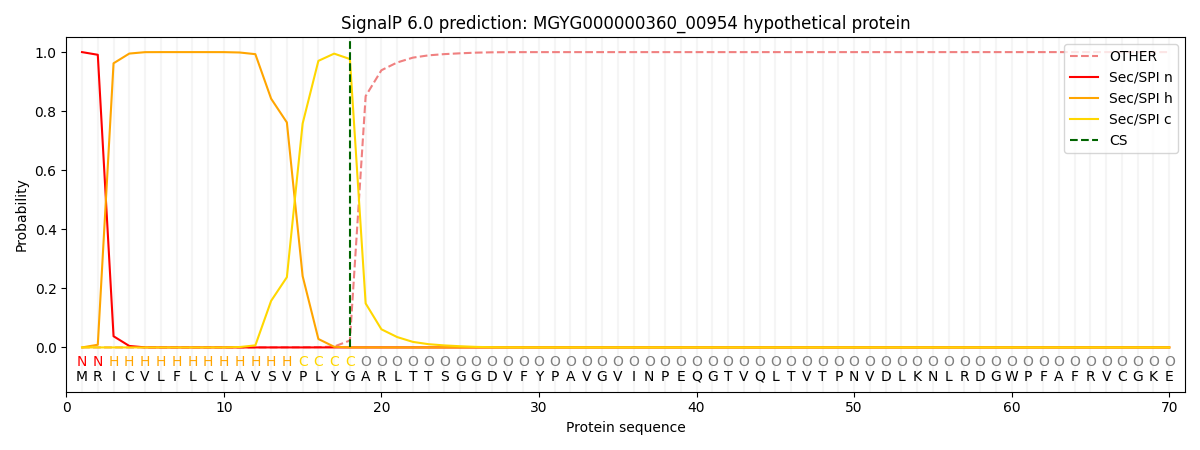
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000257 | 0.999150 | 0.000147 | 0.000146 | 0.000135 | 0.000129 |
