You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000360_02474
You are here: Home > Sequence: MGYG000000360_02474
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900548385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900548385 | |||||||||||
| CAZyme ID | MGYG000000360_02474 | |||||||||||
| CAZy Family | GH167 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 322893; End: 326819 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH167 | 522 | 934 | 3.5e-36 | 0.5648535564853556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 7.03e-11 | 526 | 727 | 100 | 295 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 6.95e-09 | 519 | 919 | 118 | 489 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46758.1 | 6.41e-122 | 278 | 1306 | 102 | 1150 |
| AVM45830.1 | 1.07e-51 | 386 | 1306 | 39 | 1014 |
| AVM45809.1 | 2.60e-41 | 1111 | 1306 | 1008 | 1214 |
| AVM44624.1 | 1.02e-40 | 1119 | 1295 | 1011 | 1186 |
| AVM43171.1 | 5.28e-40 | 1111 | 1291 | 1004 | 1184 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3TTS_A | 2.17e-06 | 499 | 679 | 93 | 273 | ChainA, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_B Chain B, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_C Chain C, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_D Chain D, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_E Chain E, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTS_F Chain F, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_A Chain A, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_B Chain B, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_C Chain C, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_D Chain D, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_E Chain E, Beta-galactosidase [Niallia circulans subsp. alkalophilus],3TTY_F Chain F, Beta-galactosidase [Niallia circulans subsp. alkalophilus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
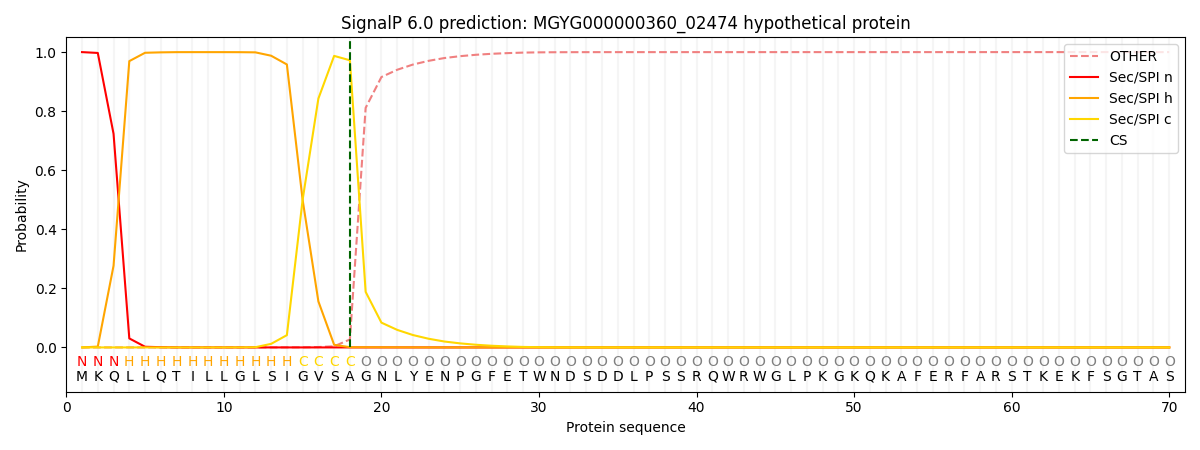
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000365 | 0.999013 | 0.000158 | 0.000147 | 0.000146 | 0.000137 |
