You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000360_03522
You are here: Home > Sequence: MGYG000000360_03522
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
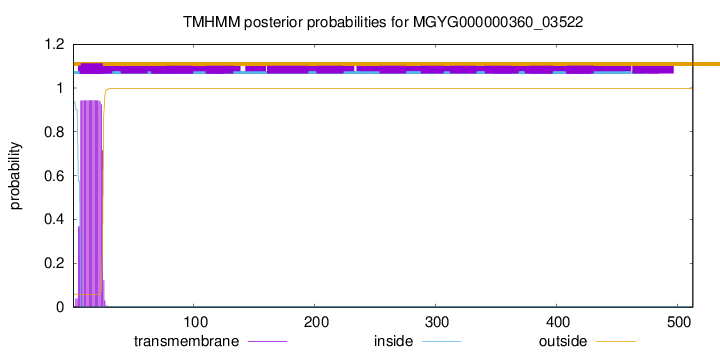
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900548385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900548385 | |||||||||||
| CAZyme ID | MGYG000000360_03522 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 77615; End: 79156 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.60e-26 | 199 | 474 | 4 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.00e-17 | 219 | 490 | 79 | 378 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM47005.1 | 2.15e-193 | 1 | 511 | 64 | 574 |
| AVM45735.1 | 6.04e-112 | 16 | 509 | 11 | 508 |
| AVM44368.1 | 8.59e-99 | 27 | 509 | 33 | 521 |
| AVM43793.1 | 3.71e-93 | 1 | 509 | 1 | 506 |
| AVM45191.1 | 1.94e-88 | 1 | 509 | 4 | 503 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NCO_A | 3.09e-17 | 305 | 476 | 119 | 295 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
| 3RJX_A | 3.09e-17 | 305 | 476 | 119 | 295 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
| 3RJY_A | 3.09e-17 | 305 | 476 | 119 | 295 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
| 6KDD_A | 8.39e-16 | 303 | 476 | 117 | 295 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 1CEC_A | 3.48e-14 | 259 | 467 | 68 | 306 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DJ77 | 1.42e-13 | 259 | 467 | 68 | 306 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P23340 | 1.42e-13 | 259 | 467 | 68 | 306 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| P0C2S3 | 1.91e-13 | 259 | 467 | 68 | 306 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| P23548 | 2.70e-10 | 220 | 449 | 84 | 323 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
| P25472 | 6.80e-09 | 219 | 448 | 62 | 269 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000508 | 0.562352 | 0.436445 | 0.000233 | 0.000227 | 0.000214 |