You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000371_00977
You are here: Home > Sequence: MGYG000000371_00977
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-390 sp003523225 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; CAG-272; CAG-390; CAG-390 sp003523225 | |||||||||||
| CAZyme ID | MGYG000000371_00977 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55170; End: 58157 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 453 | 867 | 3.3e-77 | 0.4933510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07690 | MFS_1 | 6.40e-26 | 9 | 375 | 1 | 346 | Major Facilitator Superfamily. |
| cd17325 | MFS_MdtG_SLC18_like | 1.72e-23 | 37 | 378 | 24 | 344 | bacterial MdtG-like and eukaryotic solute carrier 18 (SLC18) family of the Major Facilitator Superfamily of transporters. This family is composed of eukaryotic solute carrier 18 (SLC18) family transporters and related bacterial multidrug resistance (MDR) transporters including several proteins from Escherichia coli such as multidrug resistance protein MdtG, from Bacillus subtilis such as multidrug resistance proteins 1 (Bmr1) and 2 (Bmr2), and from Staphylococcus aureus such as quinolone resistance protein NorA. The family also includes Escherichia coli arabinose efflux transporters YfcJ and YhhS. MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. The SLC18 transporter family includes vesicular monoamine transporters (VAT1 and VAT2), vesicular acetylcholine transporter (VAChT), and SLC18B1, which is proposed to be a vesicular polyamine transporter (VPAT). The MdtG/SLC18 family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| COG3250 | LacZ | 2.48e-20 | 459 | 768 | 15 | 364 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 6.95e-20 | 458 | 849 | 14 | 443 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.73e-18 | 461 | 849 | 78 | 470 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTE67451.1 | 9.81e-171 | 441 | 982 | 15 | 553 |
| QUA53005.1 | 6.27e-169 | 441 | 976 | 15 | 547 |
| QTE72185.1 | 1.25e-167 | 441 | 983 | 15 | 554 |
| QTE73173.1 | 1.76e-167 | 441 | 983 | 15 | 554 |
| QUC67168.1 | 9.80e-167 | 441 | 983 | 15 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 1.67e-92 | 438 | 985 | 22 | 588 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6U7J_A | 3.69e-19 | 452 | 839 | 17 | 428 | UnculturedClostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_B Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_C Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.],6U7J_D Uncultured Clostridium sp. Beta-glucuronidase [uncultured Clostridium sp.] |
| 7VQM_A | 2.28e-16 | 422 | 743 | 13 | 342 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
| 6D8K_A | 1.08e-14 | 501 | 743 | 99 | 346 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 6D89_A | 2.48e-14 | 501 | 743 | 100 | 346 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P06864 | 2.05e-13 | 455 | 850 | 72 | 471 | Evolved beta-galactosidase subunit alpha OS=Escherichia coli (strain K12) OX=83333 GN=ebgA PE=1 SV=4 |
| T2KPJ7 | 6.34e-12 | 519 | 904 | 124 | 529 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| Q75W54 | 3.50e-11 | 519 | 828 | 104 | 464 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| P08236 | 3.67e-11 | 515 | 849 | 117 | 477 | Beta-glucuronidase OS=Homo sapiens OX=9606 GN=GUSB PE=1 SV=2 |
| O77695 | 8.34e-11 | 492 | 849 | 93 | 474 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000009 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
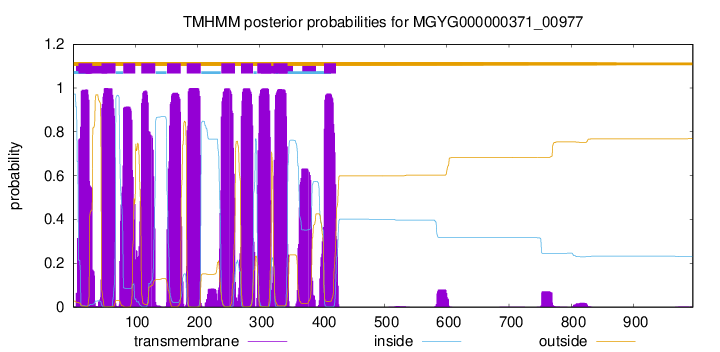
| start | end |
|---|---|
| 9 | 31 |
| 46 | 68 |
| 81 | 100 |
| 110 | 132 |
| 151 | 173 |
| 183 | 205 |
| 238 | 260 |
| 270 | 289 |
| 296 | 318 |
| 322 | 344 |
| 403 | 422 |
