You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000387_01449
You are here: Home > Sequence: MGYG000000387_01449
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000387_01449 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 193; End: 2700 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 162 | 406 | 1.3e-26 | 0.7029702970297029 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 1.16e-11 | 159 | 404 | 19 | 166 | Amb_all domain. |
| cd14256 | Dockerin_I | 7.23e-11 | 777 | 832 | 2 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| cd14253 | Dockerin | 2.46e-07 | 777 | 832 | 1 | 56 | Dockerin repeat domain. Dockerins are modules in the cellulosome complex that often anchor catalytic subunits by binding to cohesin domains of scaffolding proteins. Three types of dockerins and their corresponding cohesin have been described in the literature. This alignment models two consecutive dockerin repeats, the functional unit. |
| pfam00404 | Dockerin_1 | 1.62e-06 | 777 | 829 | 1 | 52 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam02368 | Big_2 | 1.64e-06 | 701 | 752 | 20 | 70 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL50775.1 | 6.43e-143 | 3 | 602 | 2 | 565 |
| BAV13078.1 | 7.02e-143 | 3 | 602 | 5 | 568 |
| QNF29855.1 | 1.52e-132 | 33 | 580 | 28 | 551 |
| QYR22341.1 | 2.28e-132 | 6 | 590 | 4 | 563 |
| AEI42963.1 | 3.92e-130 | 6 | 606 | 4 | 575 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 1.83e-48 | 58 | 398 | 9 | 289 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
| 5AMV_A | 3.30e-10 | 75 | 478 | 28 | 366 | Structuralinsights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
| 1BN8_A | 3.59e-10 | 75 | 478 | 49 | 387 | BacillusSubtilis Pectate Lyase [Bacillus subtilis] |
| 2BSP_A | 8.33e-10 | 75 | 478 | 49 | 387 | ChainA, PROTEIN (PECTATE LYASE) [Bacillus subtilis] |
| 2NZM_A | 1.79e-09 | 75 | 478 | 28 | 366 | ChainA, Pectate lyase [Bacillus subtilis],2O04_A Chain A, Pectate lyase [Bacillus subtilis],2O0V_A Chain A, Pectate lyase [Bacillus subtilis],2O0W_A Chain A, Pectate lyase [Bacillus subtilis],2O17_A Chain A, Pectate lyase [Bacillus subtilis],2O1D_A Chain A, Pectate lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 1.54e-48 | 39 | 476 | 20 | 385 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
| P33747 | 5.94e-13 | 583 | 753 | 24 | 192 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P39116 | 1.96e-09 | 75 | 478 | 49 | 387 | Pectate lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=pel PE=1 SV=1 |
| Q9WYR4 | 2.65e-08 | 186 | 268 | 119 | 189 | Pectate trisaccharide-lyase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=pelA PE=1 SV=1 |
| B1L969 | 6.13e-08 | 186 | 268 | 117 | 187 | Pectate trisaccharide-lyase OS=Thermotoga sp. (strain RQ2) OX=126740 GN=pelA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
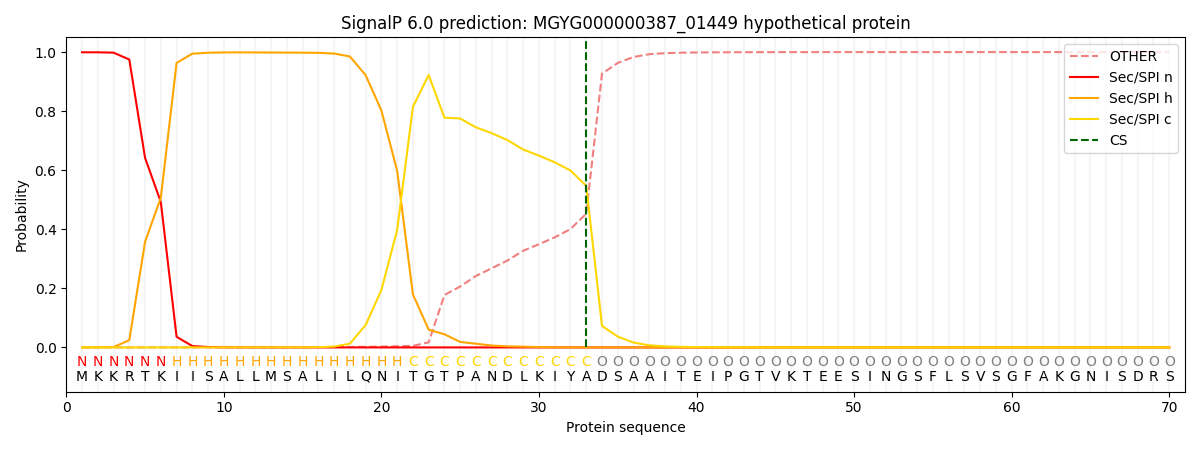
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000721 | 0.997644 | 0.001039 | 0.000211 | 0.000186 | 0.000179 |
