You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000504_01143
Basic Information
help
| Species |
CAG-312 sp000438015
|
| Lineage |
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp000438015
|
| CAZyme ID |
MGYG000000504_01143
|
| CAZy Family |
GH110 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000504 |
2519783 |
MAG |
Fiji |
Oceania |
|
| Gene Location |
Start: 33536;
End: 38053
Strand: -
|
No EC number prediction in MGYG000000504_01143.
| Family |
Start |
End |
Evalue |
family coverage |
| GH110 |
613 |
1150 |
2.4e-46 |
0.9872262773722628 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13229
|
Beta_helix |
2.49e-07 |
980 |
1151 |
28 |
157 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
1.28e-06 |
981 |
1178 |
6 |
156 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229
|
Beta_helix |
1.53e-04 |
979 |
1120 |
50 |
157 |
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
B1V8K7
|
2.43e-07 |
799 |
1119 |
260 |
582 |
Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
This protein is predicted as LIPO
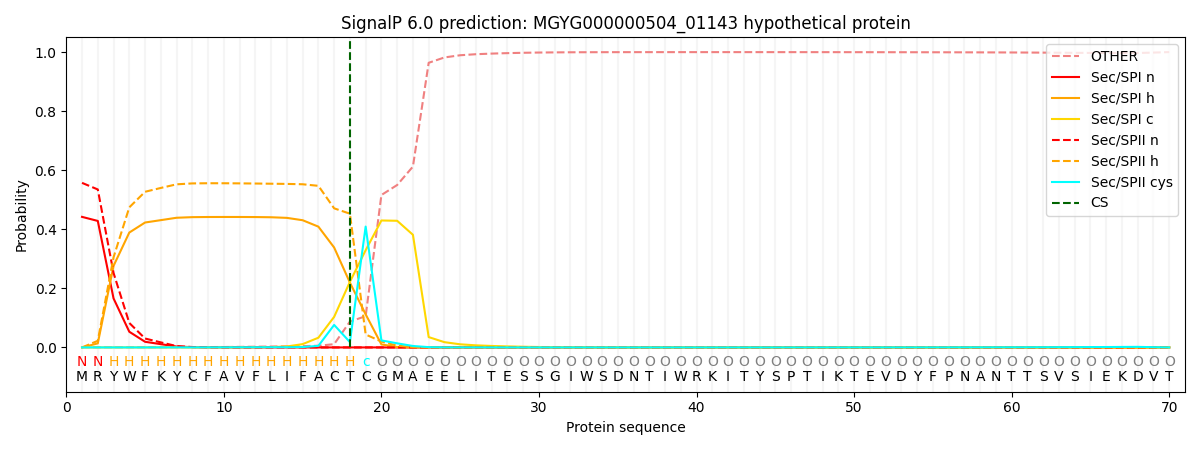
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.002238
|
0.430580
|
0.566234
|
0.000355
|
0.000299
|
0.000281
|
There is no transmembrane helices in MGYG000000504_01143.

