You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000543_00701
You are here: Home > Sequence: MGYG000000543_00701
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
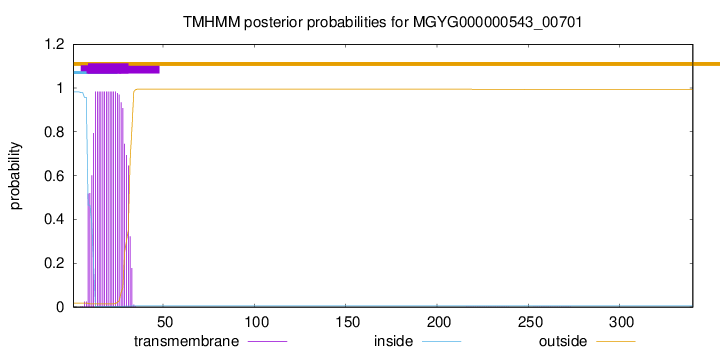
TMHMM annotations
Basic Information help
| Species | Agathobacter sp900552085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp900552085 | |||||||||||
| CAZyme ID | MGYG000000543_00701 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30964; End: 31986 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 50 | 293 | 3.4e-97 | 0.9915611814345991 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.27e-72 | 48 | 301 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.74e-09 | 67 | 301 | 77 | 363 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam16444 | DUF5041 | 0.006 | 174 | 236 | 74 | 133 | Domain of unknown function (DUF5041). This family consists of several uncharacterized proteins around 230 residues in length and is mainly found in various Bacteroidales species. The function of this family is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFA47670.1 | 2.60e-132 | 40 | 336 | 205 | 501 |
| QBR94633.1 | 2.62e-126 | 34 | 335 | 88 | 388 |
| QNM00780.1 | 2.18e-124 | 40 | 329 | 221 | 508 |
| QWT53734.1 | 7.02e-123 | 40 | 329 | 221 | 508 |
| AWW27905.1 | 1.05e-121 | 40 | 309 | 228 | 497 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 4.28e-98 | 40 | 335 | 13 | 302 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 1.22e-95 | 35 | 329 | 32 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 3.74e-91 | 35 | 329 | 7 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 4.03e-88 | 40 | 329 | 12 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 5WH8_A | 3.57e-83 | 29 | 329 | 6 | 303 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 6.79e-98 | 40 | 331 | 116 | 405 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P15704 | 1.69e-97 | 3 | 329 | 9 | 331 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 1.05e-93 | 13 | 329 | 12 | 325 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| Q07940 | 1.52e-92 | 53 | 329 | 17 | 290 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P23549 | 9.28e-92 | 13 | 329 | 12 | 325 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000362 | 0.998961 | 0.000192 | 0.000184 | 0.000157 | 0.000149 |