You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000566_00245
You are here: Home > Sequence: MGYG000000566_00245
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Mailhella; | |||||||||||
| CAZyme ID | MGYG000000566_00245 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1148; End: 3415 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 145 | 602 | 6.7e-63 | 0.8240740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 2.03e-27 | 143 | 639 | 8 | 499 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 3.65e-13 | 147 | 461 | 9 | 317 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 5.56e-12 | 198 | 359 | 1 | 157 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| pfam05466 | BASP1 | 9.99e-07 | 6 | 116 | 70 | 203 | Brain acid soluble protein 1 (BASP1 protein). This family consists of several brain acid soluble protein 1 (BASP1) or neuronal axonal membrane protein NAP-22. The BASP1 is a neuron enriched Ca(2+)-dependent calmodulin-binding protein of unknown function. |
| PHA03169 | PHA03169 | 4.18e-06 | 9 | 115 | 119 | 216 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAS95692.1 | 9.34e-134 | 133 | 708 | 2 | 579 |
| ABM28860.1 | 5.64e-133 | 133 | 708 | 65 | 642 |
| ADP86283.1 | 5.64e-133 | 133 | 708 | 65 | 642 |
| ABB39217.1 | 1.53e-100 | 122 | 699 | 27 | 605 |
| BBD06923.1 | 1.34e-81 | 134 | 709 | 6 | 559 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 1.93e-10 | 144 | 482 | 32 | 369 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4KC80 | 3.59e-20 | 145 | 486 | 7 | 341 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT1 PE=3 SV=1 |
| Q4K884 | 3.71e-18 | 128 | 517 | 21 | 405 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT2 PE=3 SV=1 |
| Q3KCC9 | 7.75e-17 | 149 | 491 | 28 | 364 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1 |
| Q02R27 | 2.23e-16 | 148 | 502 | 9 | 362 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=arnT PE=3 SV=1 |
| Q9HY61 | 2.96e-16 | 148 | 502 | 9 | 362 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000041 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
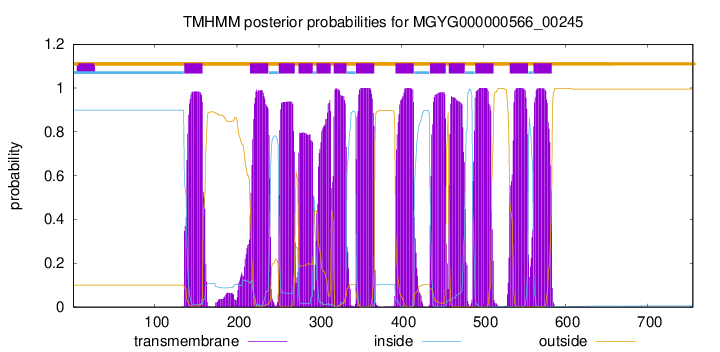
| start | end |
|---|---|
| 136 | 158 |
| 216 | 238 |
| 251 | 270 |
| 275 | 292 |
| 297 | 314 |
| 318 | 333 |
| 345 | 367 |
| 393 | 415 |
| 435 | 454 |
| 458 | 477 |
| 490 | 512 |
| 532 | 554 |
| 561 | 583 |
