You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000574_02304
You are here: Home > Sequence: MGYG000000574_02304
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000574_02304 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8; End: 3448 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 658 | 1144 | 1.6e-95 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 3.36e-08 | 598 | 648 | 7 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam13229 | Beta_helix | 6.88e-08 | 833 | 1014 | 10 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam00404 | Dockerin_1 | 2.44e-07 | 598 | 648 | 6 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam00754 | F5_F8_type_C | 1.18e-05 | 226 | 365 | 4 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13229 | Beta_helix | 4.30e-05 | 744 | 956 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA49029.1 | 1.33e-50 | 657 | 1143 | 27 | 508 |
| QTH41501.1 | 3.65e-50 | 656 | 1144 | 323 | 817 |
| BCA49028.1 | 5.47e-50 | 660 | 1133 | 42 | 508 |
| QHT63103.1 | 1.29e-49 | 656 | 1143 | 34 | 518 |
| QHW32944.1 | 9.48e-49 | 656 | 1143 | 34 | 518 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KQT_A | 9.64e-32 | 656 | 1043 | 242 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 9.31e-31 | 656 | 1043 | 242 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 7V6M_A | 2.11e-27 | 658 | 1143 | 8 | 574 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
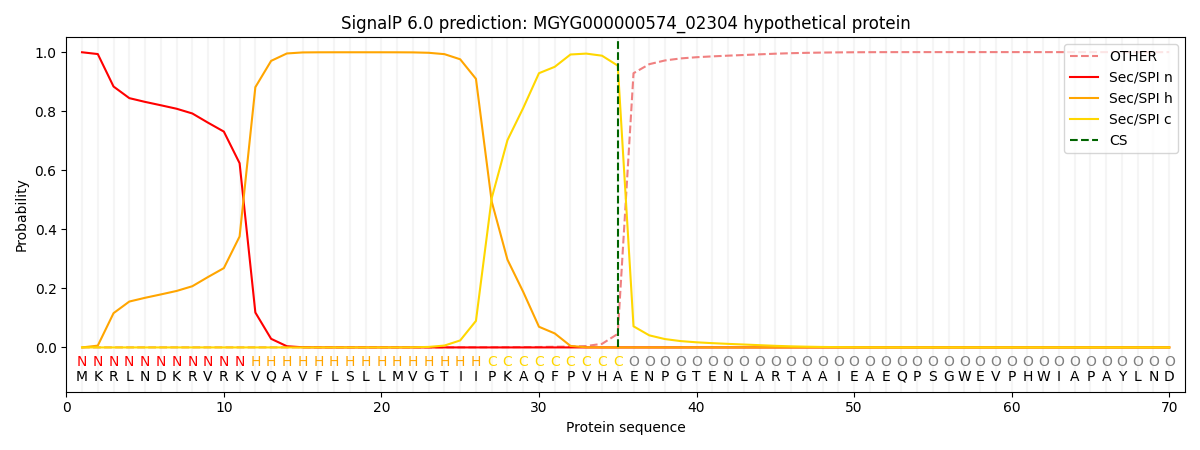
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000965 | 0.997704 | 0.000643 | 0.000282 | 0.000199 | 0.000171 |
