You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000602_00486
You are here: Home > Sequence: MGYG000000602_00486
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-1427 sp900544455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Eggerthellaceae; CAG-1427; CAG-1427 sp900544455 | |||||||||||
| CAZyme ID | MGYG000000602_00486 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20159; End: 21478 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 52 | 278 | 5.6e-29 | 0.9826086956521739 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06438 | EpsO_like | 6.28e-80 | 55 | 240 | 1 | 183 | EpsO protein participates in the methanolan synthesis. The Methylobacillus sp EpsO protein is predicted to participate in the methanolan synthesis. Methanolan is an exopolysaccharide (EPS), composed of glucose, mannose and galactose. A 21 genes cluster was predicted to participate in the methanolan synthesis. Gene disruption analysis revealed that EpsO is one of the glycosyltransferase enzymes involved in the synthesis of repeating sugar units onto the lipid carrier. |
| COG1215 | BcsA | 1.41e-43 | 8 | 406 | 11 | 401 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| cd06423 | CESA_like | 1.97e-37 | 55 | 237 | 1 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| pfam13641 | Glyco_tranf_2_3 | 5.11e-25 | 52 | 280 | 3 | 230 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| PRK11204 | PRK11204 | 1.21e-16 | 31 | 366 | 31 | 363 | N-glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAK44311.1 | 1.38e-250 | 1 | 433 | 1 | 433 |
| ACV55180.1 | 6.35e-245 | 1 | 431 | 1 | 431 |
| QOS69441.1 | 3.00e-243 | 1 | 431 | 1 | 431 |
| QTU83730.1 | 9.89e-239 | 1 | 435 | 1 | 435 |
| BAN76823.1 | 1.96e-229 | 7 | 431 | 10 | 434 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P74165 | 8.19e-18 | 42 | 304 | 95 | 355 | Beta-monoglucosyldiacylglycerol synthase OS=Synechocystis sp. (strain PCC 6803 / Kazusa) OX=1111708 GN=sll1377 PE=1 SV=1 |
| Q5HCN1 | 1.13e-15 | 4 | 311 | 2 | 294 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain COL) OX=93062 GN=icaA PE=3 SV=1 |
| Q6GDD8 | 1.13e-15 | 4 | 311 | 2 | 294 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=icaA PE=3 SV=1 |
| Q7A351 | 1.13e-15 | 4 | 311 | 2 | 294 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain N315) OX=158879 GN=icaA PE=3 SV=1 |
| Q99QX3 | 1.13e-15 | 4 | 311 | 2 | 294 | Poly-beta-1,6-N-acetyl-D-glucosamine synthase OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=icaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
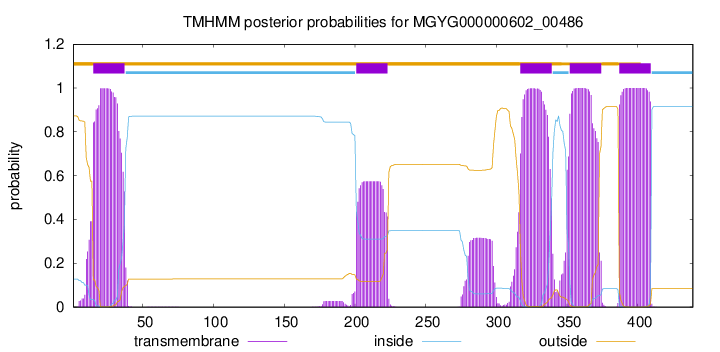
| start | end |
|---|---|
| 15 | 37 |
| 201 | 223 |
| 317 | 339 |
| 352 | 374 |
| 387 | 409 |
