You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000642_00110
You are here: Home > Sequence: MGYG000000642_00110
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
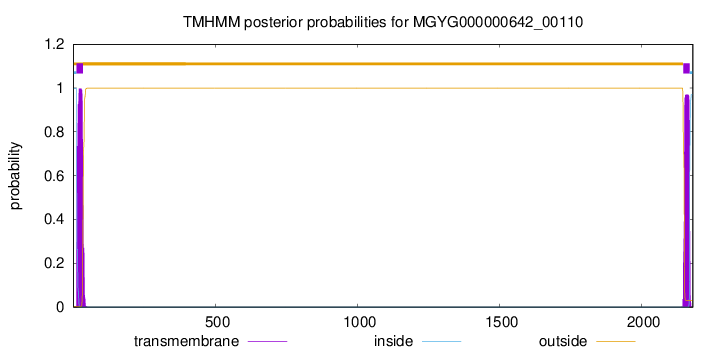
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Arcanobacterium_A; | |||||||||||
| CAZyme ID | MGYG000000642_00110 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 124963; End: 131505 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 926 | 1494 | 5.6e-148 | 0.9938900203665988 |
| GH33 | 428 | 800 | 9.2e-73 | 0.9181286549707602 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.64e-76 | 435 | 812 | 15 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 1.48e-32 | 423 | 813 | 261 | 659 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 5.96e-17 | 445 | 794 | 1 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam18957 | RibLong | 1.64e-11 | 1994 | 2080 | 6 | 93 | Long Rib domain. This entry represents the Long Rib domain that is closely related to the pfam08428 Rib domain but has a conserved insertion. These domains are found in bacterial cell surface proteins. |
| pfam18957 | RibLong | 1.83e-07 | 1786 | 1880 | 14 | 92 | Long Rib domain. This entry represents the Long Rib domain that is closely related to the pfam08428 Rib domain but has a conserved insertion. These domains are found in bacterial cell surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ESV34046.1 | 1.14e-201 | 909 | 1890 | 29 | 1064 |
| QSG89108.1 | 1.27e-200 | 909 | 1969 | 29 | 1155 |
| VEG79925.1 | 1.27e-200 | 909 | 1969 | 29 | 1155 |
| BAJ67172.1 | 1.27e-200 | 909 | 1969 | 29 | 1155 |
| BBV24031.1 | 1.57e-200 | 909 | 1890 | 29 | 1064 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 1.21e-180 | 924 | 1499 | 6 | 580 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 2.82e-157 | 911 | 1497 | 2 | 598 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 5.26e-151 | 926 | 1501 | 13 | 614 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 9.45e-113 | 926 | 1348 | 244 | 660 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 1.67e-108 | 926 | 1348 | 244 | 660 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 1.31e-82 | 433 | 818 | 66 | 403 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P29767 | 5.64e-17 | 432 | 816 | 393 | 826 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| Q5RAF4 | 7.79e-15 | 430 | 798 | 73 | 387 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
| Q99519 | 7.79e-15 | 430 | 798 | 73 | 387 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
| A6BMK7 | 1.38e-14 | 430 | 768 | 73 | 356 | Sialidase-1 OS=Bos taurus OX=9913 GN=NEU1 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
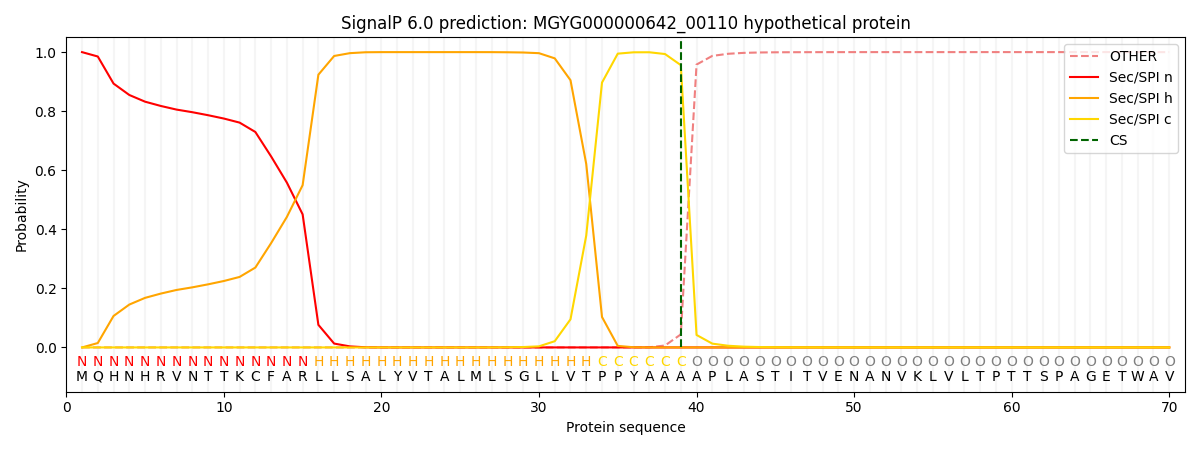
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000466 | 0.998706 | 0.000225 | 0.000234 | 0.000175 | 0.000154 |