You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000642_00587
You are here: Home > Sequence: MGYG000000642_00587
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
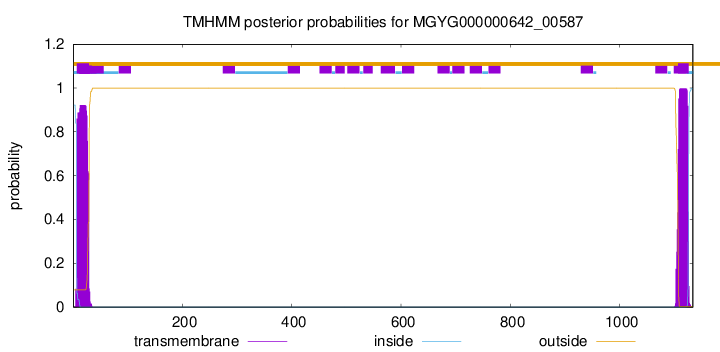
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Arcanobacterium_A; | |||||||||||
| CAZyme ID | MGYG000000642_00587 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 159711; End: 163115 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 201 | 754 | 1.2e-157 | 0.9938900203665988 |
| CBM32 | 45 | 192 | 3.7e-16 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.61e-11 | 41 | 166 | 1 | 106 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam05345 | He_PIG | 2.15e-07 | 952 | 1049 | 1 | 94 | Putative Ig domain. This alignment represents the conserved core region of ~90 residue repeat found in several haemagglutinins and other cell surface proteins. Sequence similarities to (pfam02494) and (pfam00801) suggest an Ig-like fold (personal obs:C. Yeats). So this family may be similar in function to the (pfam02639) and (pfam02638) domains. This domain is also found in the WisP family of proteins of Tropheryma whipplei. |
| smart00736 | CADG | 1.57e-04 | 997 | 1051 | 35 | 93 | Dystroglycan-type cadherin-like domains. Cadherin-homologous domains present in metazoan dystroglycans and alpha/epsilon sarcoglycans, yeast Axl2p and in a very large protein from magnetotactic bacteria. Likely to bind calcium ions. |
| cd00057 | FA58C | 0.001 | 32 | 119 | 1 | 85 | Substituted updates: Jan 31, 2002 |
| pfam13229 | Beta_helix | 0.005 | 401 | 527 | 5 | 117 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADP35567.1 | 1.29e-177 | 186 | 969 | 52 | 931 |
| QTQ17888.1 | 1.36e-174 | 186 | 1048 | 18 | 993 |
| VEG19581.1 | 1.96e-173 | 186 | 1048 | 18 | 993 |
| BAQ97658.1 | 1.96e-173 | 186 | 1048 | 18 | 993 |
| ADO52688.1 | 2.08e-167 | 186 | 1048 | 18 | 991 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 3.28e-178 | 198 | 753 | 5 | 574 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 1.82e-142 | 187 | 755 | 3 | 596 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 2.39e-133 | 194 | 755 | 6 | 608 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 1.64e-114 | 190 | 607 | 233 | 644 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 1.72e-110 | 190 | 617 | 233 | 653 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 6.59e-08 | 42 | 175 | 631 | 746 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 6.59e-08 | 42 | 175 | 631 | 746 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| B2UL12 | 1.82e-06 | 204 | 267 | 39 | 101 | Alpha-1,3-galactosidase A OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
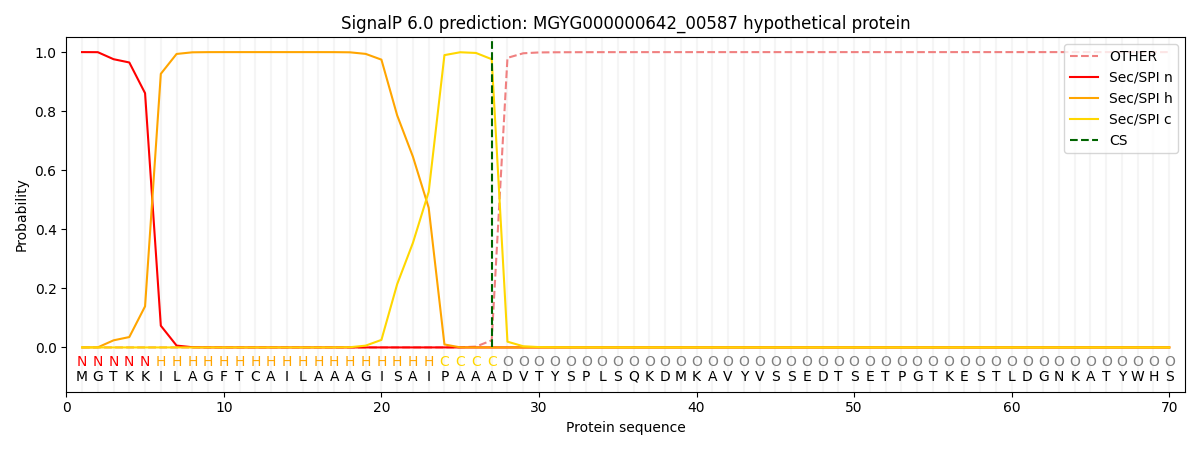
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998946 | 0.000171 | 0.000207 | 0.000185 | 0.000160 |