You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000667_01233
Basic Information
help
| Species |
UBA7173 sp900546835
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; UBA7173 sp900546835
|
| CAZyme ID |
MGYG000000667_01233
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 581 |
|
65789.07 |
8.0007 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000667 |
2502097 |
MAG |
Kazakhstan |
Asia |
|
| Gene Location |
Start: 58203;
End: 59948
Strand: -
|
No EC number prediction in MGYG000000667_01233.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
5 |
480 |
5.9e-74 |
0.8296296296296296 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
5.28e-27 |
6 |
385 |
6 |
370 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
4.17e-06 |
64 |
225 |
2 |
160 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O67270
|
2.52e-15 |
10 |
353 |
5 |
321 |
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
|
A8FRR0
|
2.46e-08 |
25 |
187 |
24 |
188 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.660417
|
0.332559
|
0.003380
|
0.001232
|
0.000680
|
0.001739
|
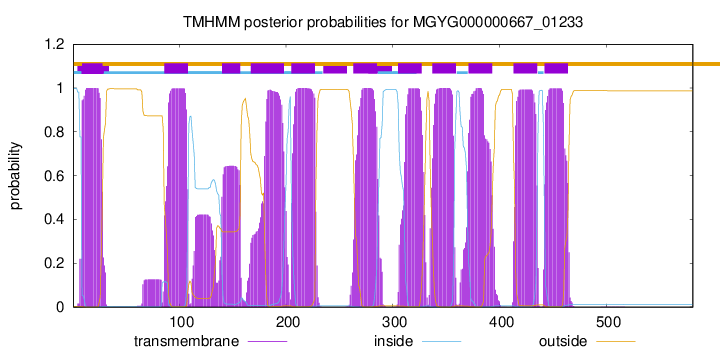
| start |
end |
| 9 |
28 |
| 86 |
108 |
| 140 |
157 |
| 167 |
198 |
| 205 |
227 |
| 263 |
285 |
| 305 |
327 |
| 337 |
359 |
| 371 |
393 |
| 413 |
435 |
| 442 |
464 |


