You are browsing environment: HUMAN GUT
MGYG000000696_00082
Basic Information
help
Species
Parabacteroides sp900549585
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900549585
CAZyme ID
MGYG000000696_00082
CAZy Family
GH163
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000696
3184747
MAG
Kazakhstan
Asia
Gene Location
Start: 97904;
End: 100111
Strand: +
No EC number prediction in MGYG000000696_00082.
Family
Start
End
Evalue
family coverage
GH163
198
451
5.2e-104
0.9880478087649402
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16126
DUF4838
2.15e-132
190
451
1
262
Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown.
more
pfam02838
Glyco_hydro_20b
9.89e-08
43
121
29
105
Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold.
more
pfam00754
F5_F8_type_C
3.18e-06
593
708
9
112
F5/8 type C domain. This domain is also known as the discoidin (DS) domain family.
more
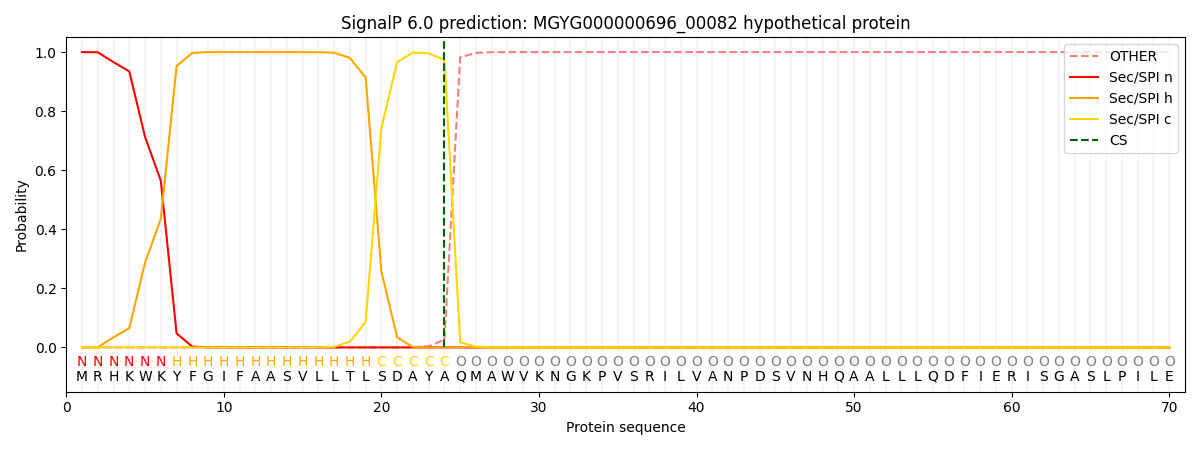
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000567
0.998662
0.000221
0.000178
0.000179
0.000168