You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000708_00046
You are here: Home > Sequence: MGYG000000708_00046
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
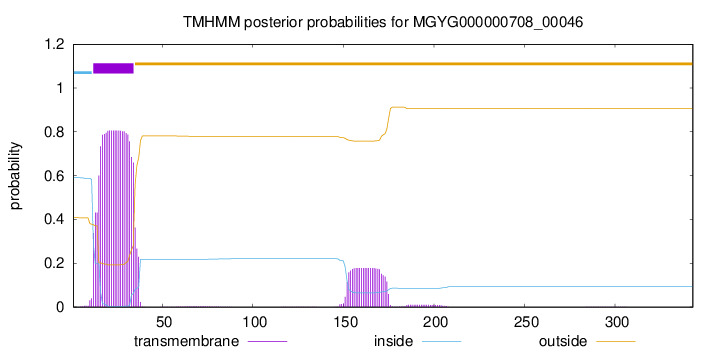
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Succinivibrionaceae; Succinivibrio; | |||||||||||
| CAZyme ID | MGYG000000708_00046 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | Minor endoglucanase Y | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9523; End: 10554 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 1.06e-77 | 30 | 338 | 1 | 318 | Glycosyl hydrolases family 8. |
| PRK11097 | PRK11097 | 4.70e-53 | 17 | 294 | 3 | 294 | cellulase. |
| COG3405 | BcsZ | 1.86e-45 | 15 | 294 | 1 | 292 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
| COG4225 | YesR | 0.003 | 68 | 130 | 140 | 211 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIX95850.1 | 2.08e-107 | 12 | 338 | 1 | 329 |
| QCZ26633.1 | 4.49e-106 | 35 | 334 | 24 | 324 |
| QIK15535.1 | 9.01e-106 | 36 | 334 | 25 | 324 |
| QJT83151.1 | 9.63e-106 | 12 | 338 | 1 | 328 |
| ANI80779.1 | 1.36e-105 | 12 | 338 | 1 | 328 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CZL_A | 1.14e-99 | 35 | 338 | 30 | 333 | ChainA, Glucanase [Raoultella ornithinolytica] |
| 5GY3_A | 7.81e-96 | 35 | 338 | 2 | 305 | ChainA, Glucanase [Klebsiella pneumoniae] |
| 6VC5_A | 1.70e-63 | 32 | 338 | 3 | 312 | 1.6Angstrom Resolution Crystal Structure of endoglucanase from Komagataeibacter sucrofermentans [Komagataeibacter sucrofermentans] |
| 1WZZ_A | 1.08e-61 | 32 | 338 | 18 | 327 | Structureof endo-beta-1,4-glucanase CMCax from Acetobacter xylinum [Komagataeibacter xylinus] |
| 4Q2B_A | 1.24e-33 | 36 | 258 | 6 | 230 | Thecrystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_B The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_C The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_D The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_E The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440],4Q2B_F The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 [Pseudomonas putida KT2440] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27032 | 2.08e-90 | 25 | 338 | 15 | 329 | Minor endoglucanase Y OS=Dickeya dadantii (strain 3937) OX=198628 GN=celY PE=1 SV=1 |
| P18336 | 1.60e-85 | 13 | 313 | 3 | 303 | Endoglucanase OS=Cellulomonas uda OX=1714 PE=1 SV=1 |
| P37696 | 3.35e-62 | 32 | 338 | 26 | 335 | Probable endoglucanase OS=Komagataeibacter hansenii OX=436 GN=cmcAX PE=1 SV=1 |
| P58935 | 1.28e-33 | 37 | 266 | 36 | 268 | Endoglucanase OS=Xanthomonas axonopodis pv. citri (strain 306) OX=190486 GN=bcsZ PE=3 SV=1 |
| Q8X5L9 | 9.34e-30 | 13 | 333 | 4 | 338 | Endoglucanase OS=Escherichia coli O157:H7 OX=83334 GN=bcsZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000552 | 0.998650 | 0.000215 | 0.000204 | 0.000184 | 0.000166 |