You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000760_00297
You are here: Home > Sequence: MGYG000000760_00297
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
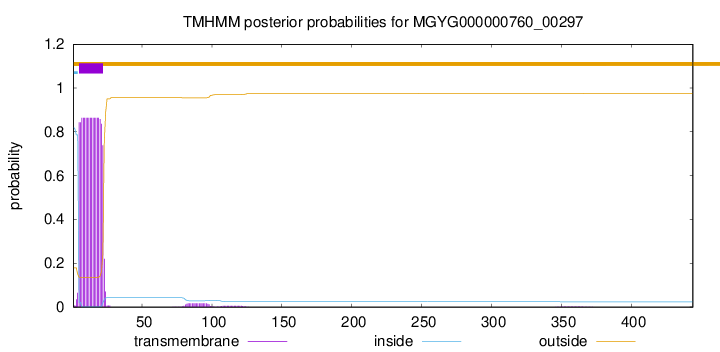
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; | |||||||||||
| CAZyme ID | MGYG000000760_00297 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 310108; End: 311442 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 77 | 393 | 8.3e-46 | 0.966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 7.50e-38 | 121 | 391 | 3 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 2.94e-35 | 101 | 395 | 24 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 6.00e-32 | 121 | 394 | 69 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 0.004 | 180 | 214 | 137 | 170 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEW21052.1 | 4.73e-182 | 32 | 436 | 25 | 427 |
| BAR51127.1 | 4.73e-182 | 32 | 436 | 25 | 427 |
| BAR48814.1 | 4.73e-182 | 32 | 436 | 25 | 427 |
| QGA25900.1 | 1.91e-172 | 26 | 435 | 23 | 431 |
| QGA28189.1 | 2.67e-117 | 8 | 434 | 3 | 494 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 2.16e-18 | 42 | 425 | 41 | 393 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1I1W_A | 5.31e-18 | 121 | 397 | 51 | 301 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
| 2BNJ_A | 9.75e-18 | 121 | 397 | 51 | 301 | Thexylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
| 1K6A_A | 4.43e-17 | 121 | 397 | 51 | 301 | Structuralstudies on the mobility in the active site of the Thermoascus aurantiacus xylanase I [Thermoascus aurantiacus] |
| 1GOK_A | 4.43e-17 | 121 | 397 | 51 | 301 | Thermostablexylanase I from Thermoascus aurantiacus- Crystal form II [Thermoascus aurantiacus],1GOM_A Thermostable xylanase I from Thermoascus aurantiacus- Crystal form I [Thermoascus aurantiacus],1GOO_A Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex [Thermoascus aurantiacus],1GOQ_A Thermostable xylanase I from Thermoascus aurantiacus - Room temperature xylobiose complex [Thermoascus aurantiacus],1GOR_A THERMOSTABLE XYLANASE I FROM THERMOASCUS AURANTIACUS - XYLOBIOSE COMPLEX AT 100 K [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q60041 | 1.60e-16 | 121 | 403 | 72 | 344 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
| O80596 | 2.23e-16 | 54 | 425 | 691 | 1033 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| P23360 | 3.30e-16 | 121 | 397 | 77 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
| O69231 | 2.20e-14 | 88 | 367 | 14 | 295 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| I1S3C6 | 3.23e-14 | 85 | 399 | 92 | 381 | Endo-1,4-beta-xylanase D OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
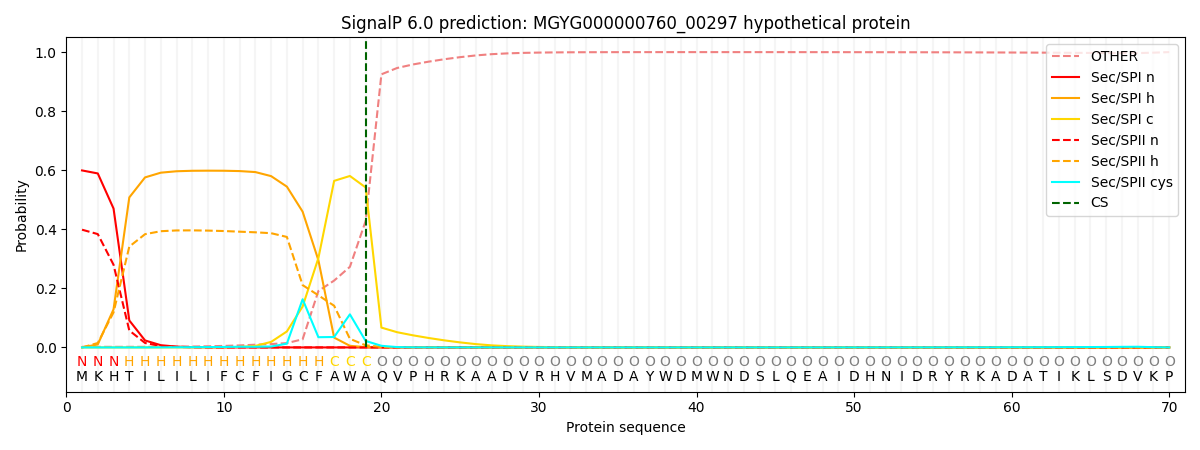
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004188 | 0.585933 | 0.408784 | 0.000339 | 0.000363 | 0.000381 |