You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000959_01028
You are here: Home > Sequence: MGYG000000959_01028
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
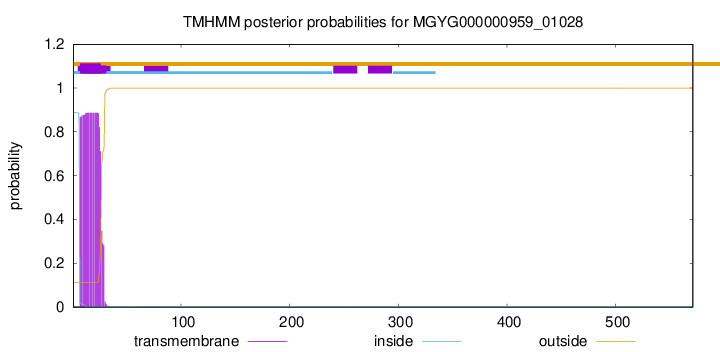
TMHMM annotations
Basic Information help
| Species | Ruminococcus sp900752785 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus; Ruminococcus sp900752785 | |||||||||||
| CAZyme ID | MGYG000000959_01028 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 167; End: 1882 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 54 | 400 | 4.1e-115 | 0.9967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 8.26e-43 | 53 | 395 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.92e-29 | 47 | 402 | 32 | 370 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd14256 | Dockerin_I | 7.54e-06 | 503 | 561 | 2 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAT48117.1 | 5.51e-270 | 1 | 489 | 1 | 497 |
| ADU21575.1 | 3.88e-205 | 10 | 556 | 12 | 560 |
| CBL16847.1 | 6.32e-198 | 34 | 519 | 36 | 516 |
| ADU73502.1 | 2.10e-181 | 1 | 515 | 1 | 514 |
| ALX07424.1 | 2.10e-181 | 1 | 515 | 1 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VRX_A | 2.97e-68 | 41 | 404 | 2 | 334 | ChainA, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
| 1ECE_A | 5.84e-68 | 41 | 404 | 2 | 334 | AcidothermusCellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
| 3W6M_A | 6.43e-47 | 44 | 404 | 7 | 359 | Contributionof disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 3VVG_A | 6.43e-47 | 44 | 404 | 7 | 359 | TheCrystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 2ZUM_A | 3.64e-46 | 44 | 404 | 40 | 392 | FunctionalAnalysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_A Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_B Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_C Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05332 | 4.19e-182 | 1 | 515 | 1 | 514 | Endoglucanase G OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celG PE=3 SV=1 |
| P10474 | 3.23e-132 | 38 | 461 | 623 | 1027 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P04956 | 1.00e-126 | 5 | 462 | 2 | 474 | Endoglucanase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celB PE=3 SV=1 |
| P50400 | 4.85e-117 | 13 | 459 | 22 | 443 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
| P23548 | 2.19e-69 | 44 | 404 | 38 | 365 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001686 | 0.990024 | 0.007359 | 0.000378 | 0.000275 | 0.000257 |