You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001000_01353
You are here: Home > Sequence: MGYG000001000_01353
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA4716 sp900556575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; UBA4716; UBA4716 sp900556575 | |||||||||||
| CAZyme ID | MGYG000001000_01353 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Xylan 1,4-beta-xylosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2042; End: 3589 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 73 | 324 | 3.9e-60 | 0.9814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03080 | PLN03080 | 2.05e-62 | 39 | 446 | 41 | 459 | Probable beta-xylosidase; Provisional |
| COG1472 | BglX | 1.11e-52 | 71 | 446 | 52 | 388 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK15098 | PRK15098 | 4.53e-44 | 12 | 443 | 7 | 448 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 2.07e-37 | 89 | 350 | 75 | 310 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam01915 | Glyco_hydro_3_C | 7.81e-10 | 395 | 465 | 2 | 77 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18582.1 | 1.09e-239 | 3 | 480 | 4 | 503 |
| AUO19701.1 | 2.32e-175 | 22 | 480 | 16 | 476 |
| AUO19977.1 | 3.92e-162 | 46 | 480 | 230 | 666 |
| QUT75533.1 | 1.50e-109 | 39 | 468 | 41 | 444 |
| AEE97207.1 | 8.66e-98 | 40 | 445 | 13 | 412 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7VC7_A | 5.96e-55 | 50 | 442 | 25 | 422 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 7VC6_A | 5.96e-55 | 50 | 442 | 25 | 422 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
| 6Q7I_A | 1.80e-51 | 39 | 440 | 41 | 441 | GH3exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7I_B GH3 exo-beta-xylosidase (XlnD) [Aspergillus nidulans FGSC A4],6Q7J_A Chain A, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4],6Q7J_B Chain B, Exo-1,4-beta-xylosidase xlnD [Aspergillus nidulans FGSC A4] |
| 5A7M_A | 1.01e-50 | 42 | 495 | 44 | 493 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5A7M_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
| 5AE6_A | 1.02e-50 | 42 | 495 | 44 | 493 | ChainA, BETA-XYLOSIDASE [Trichoderma reesei],5AE6_B Chain B, BETA-XYLOSIDASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 1.02e-67 | 38 | 448 | 22 | 432 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| A5JTQ2 | 1.07e-63 | 42 | 443 | 56 | 462 | Beta-xylosidase/alpha-L-arabinofuranosidase 1 (Fragment) OS=Medicago sativa subsp. varia OX=36902 GN=Xyl1 PE=1 SV=1 |
| Q0CB82 | 3.35e-62 | 42 | 429 | 44 | 434 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=bxlB PE=3 SV=1 |
| B8NYD8 | 1.41e-61 | 42 | 431 | 42 | 434 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bxlB PE=3 SV=1 |
| Q2TYT2 | 2.53e-61 | 42 | 431 | 63 | 455 | Probable exo-1,4-beta-xylosidase bxlB OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
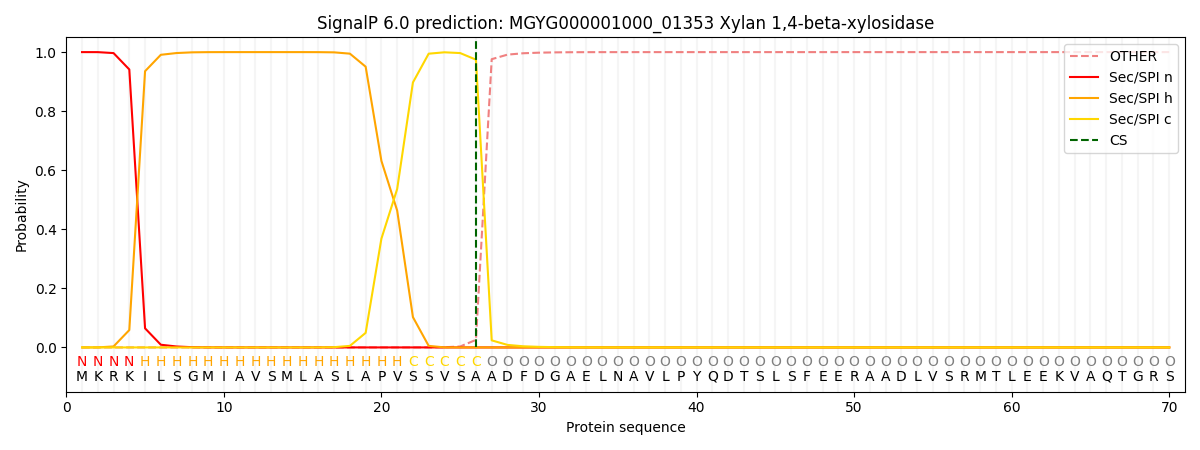
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000234 | 0.999072 | 0.000166 | 0.000185 | 0.000166 | 0.000145 |
