You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001030_02638
You are here: Home > Sequence: MGYG000001030_02638
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900760735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900760735 | |||||||||||
| CAZyme ID | MGYG000001030_02638 | |||||||||||
| CAZy Family | GH32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30743; End: 32272 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH32 | 52 | 339 | 1.2e-42 | 0.9419795221843004 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08995 | GH32_EcAec43-like | 1.90e-113 | 60 | 348 | 1 | 281 | Glycosyl hydrolase family 32, such as the putative glycoside hydrolase Escherichia coli Aec43 (FosGH2). This glycosyl hydrolase family 32 (GH32) subgroup includes Escherichia coli strain BEN2908 putative glycoside hydrolase Aec43 (FosGH2). GH32 enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). GH32 family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. |
| pfam16346 | DUF4975 | 8.53e-55 | 346 | 506 | 1 | 176 | Domain of unknown function (DUF4975). This family consists of uncharacterized proteins around 500 residues in length and is mainly found in various Bacteroides species. Several proteins in this family are annotated as Glycosyl hydrolases, but the function of this protein is unknown. |
| cd08996 | GH32_FFase | 2.68e-32 | 124 | 335 | 65 | 273 | Glycosyl hydrolase family 32, beta-fructosidases. Glycosyl hydrolase family GH32 cleaves sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| smart00640 | Glyco_32 | 6.86e-27 | 54 | 474 | 4 | 436 | Glycosyl hydrolases family 32. |
| cd18609 | GH32-like | 4.35e-26 | 95 | 327 | 48 | 295 | Glycosyl hydrolase family 32 family protein. The GH32 family contains glycosyl hydrolase family GH32 proteins that cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). This family also contains other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEA21332.1 | 1.39e-219 | 1 | 506 | 1 | 528 |
| QUB88319.1 | 1.39e-219 | 1 | 506 | 1 | 528 |
| QUB90812.1 | 1.97e-219 | 1 | 506 | 1 | 528 |
| QUB83695.1 | 5.61e-219 | 1 | 506 | 1 | 528 |
| QUI94038.1 | 4.57e-218 | 1 | 506 | 1 | 528 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6R3R_A | 3.06e-182 | 26 | 508 | 6 | 501 | Firstcrystal structure of endo-levanase BT1760 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron] |
| 6R3U_A | 2.48e-181 | 26 | 508 | 6 | 501 | Endo-levanaseBT1760 mutant E221A from Bacteroides thetaiotaomicron complexed with levantetraose [Bacteroides thetaiotaomicron] |
| 7VCO_A | 7.59e-09 | 124 | 486 | 100 | 461 | ChainA, Sucrose-6-phosphate hydrolase [Frischella perrara],7VCP_A Chain A, Sucrose-6-phosphate hydrolase [Frischella perrara] |
| 7BWB_A | 1.32e-08 | 43 | 359 | 45 | 356 | Bombyxmori GH32 beta-fructofuranosidase BmSUC1 [Bombyx mori] |
| 1UYP_A | 1.12e-07 | 47 | 359 | 3 | 301 | Thethree-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8],1UYP_B The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8],1UYP_C The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8],1UYP_D The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8],1UYP_E The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8],1UYP_F The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05936 | 2.68e-10 | 31 | 395 | 19 | 380 | Sucrose-6-phosphate hydrolase OS=Staphylococcus xylosus OX=1288 GN=scrB PE=3 SV=1 |
| O33833 | 2.00e-07 | 47 | 359 | 3 | 301 | Beta-fructosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=bfrA PE=1 SV=1 |
| P16553 | 2.88e-07 | 93 | 407 | 67 | 384 | Raffinose invertase OS=Escherichia coli OX=562 GN=rafD PE=3 SV=1 |
| P40714 | 6.69e-07 | 93 | 407 | 68 | 385 | Sucrose-6-phosphate hydrolase OS=Escherichia coli OX=562 GN=cscA PE=3 SV=1 |
| Q01IS8 | 5.34e-06 | 51 | 346 | 51 | 370 | Beta-fructofuranosidase, insoluble isoenzyme 3 OS=Oryza sativa subsp. indica OX=39946 GN=CIN3 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
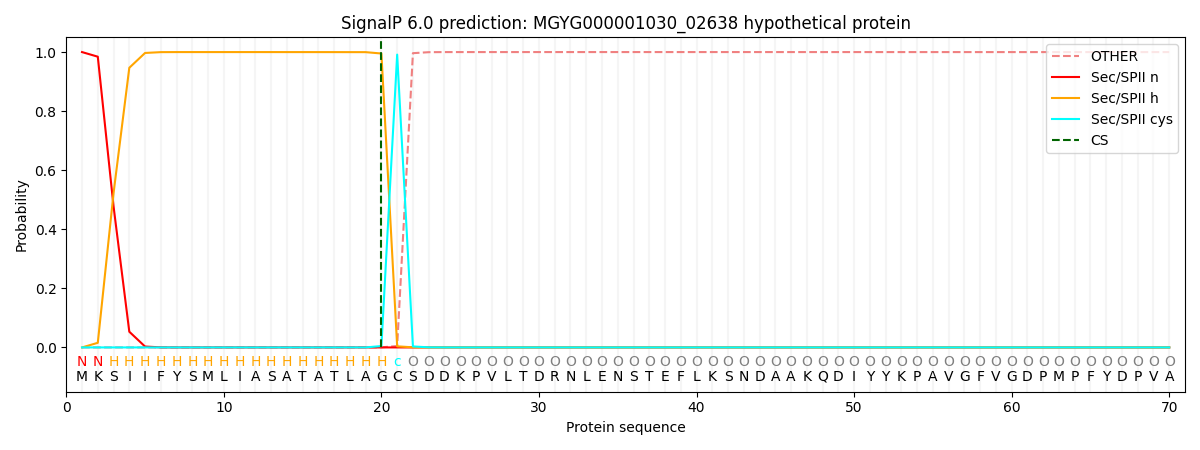
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000075 | 0.000000 | 0.000000 | 0.000000 |
