You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001134_01505
You are here: Home > Sequence: MGYG000001134_01505
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
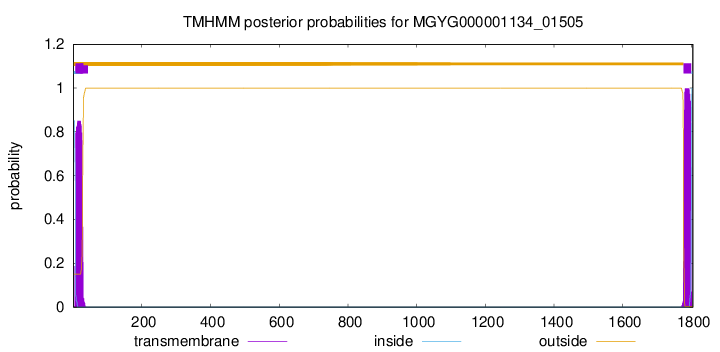
TMHMM annotations
Basic Information help
| Species | Collinsella sp900757495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; Collinsella sp900757495 | |||||||||||
| CAZyme ID | MGYG000001134_01505 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26438; End: 31852 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 114 | 235 | 5.7e-20 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.05e-16 | 108 | 244 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam09479 | Flg_new | 3.54e-15 | 1664 | 1726 | 2 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| NF033189 | internalin_A | 2.22e-07 | 1643 | 1801 | 617 | 794 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| pfam13385 | Laminin_G_3 | 3.92e-07 | 1220 | 1337 | 17 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd00057 | FA58C | 6.65e-06 | 114 | 230 | 17 | 134 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCC62414.1 | 0.0 | 257 | 1735 | 26 | 1571 |
| QQA11075.1 | 2.19e-187 | 236 | 1299 | 92 | 1166 |
| AOY54034.1 | 5.83e-187 | 236 | 1299 | 92 | 1166 |
| BAB80970.1 | 8.07e-187 | 236 | 1299 | 92 | 1166 |
| ABG83191.1 | 1.55e-186 | 236 | 1299 | 92 | 1166 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2J7M_A | 8.96e-09 | 114 | 246 | 17 | 147 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 9.16e-09 | 114 | 246 | 18 | 148 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 2.15e-07 | 114 | 246 | 605 | 735 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
| 2Y5P_A | 3.34e-06 | 1664 | 1725 | 6 | 68 | B-repeatof Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_B B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_C B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD],2Y5P_D B-repeat of Listeria monocytogenes InlB (internalin B) [Listeria monocytogenes EGD] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XL08 | 7.68e-07 | 114 | 258 | 635 | 772 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 1.32e-06 | 114 | 246 | 635 | 765 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
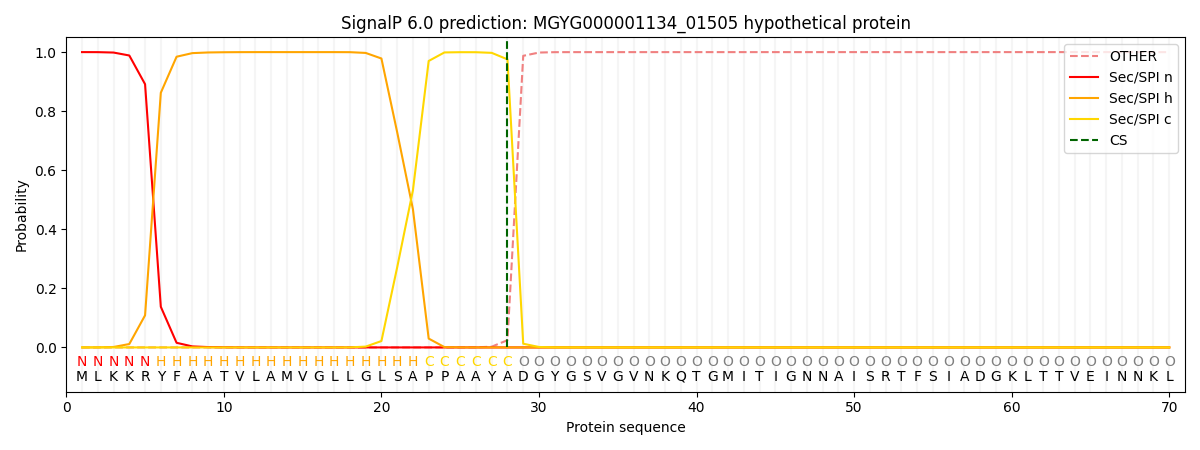
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000391 | 0.998688 | 0.000215 | 0.000252 | 0.000222 | 0.000194 |