You are browsing environment: HUMAN GUT
MGYG000001239_01303
Basic Information
help
Species
CAG-776 sp900758895
Lineage
Bacteria; Firmicutes; Bacilli; RF39; UBA660; CAG-776; CAG-776 sp900758895
CAZyme ID
MGYG000001239_01303
CAZy Family
GH105
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
379
44186.07
8.7378
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001239
1436828
MAG
Austria
Europe
Gene Location
Start: 709;
End: 1848
Strand: -
No EC number prediction in MGYG000001239_01303.
Family
Start
End
Evalue
family coverage
GH105
138
350
1.4e-23
0.6807228915662651
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam07470
Glyco_hydro_88
4.84e-10
180
334
128
291
Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases.
more
COG4225
YesR
0.004
175
340
129
309
Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism].
more
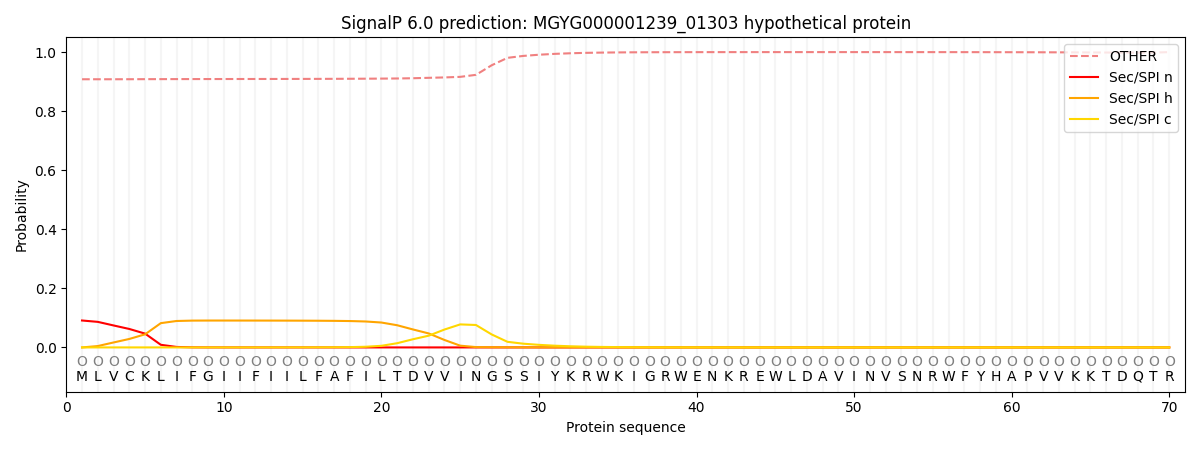
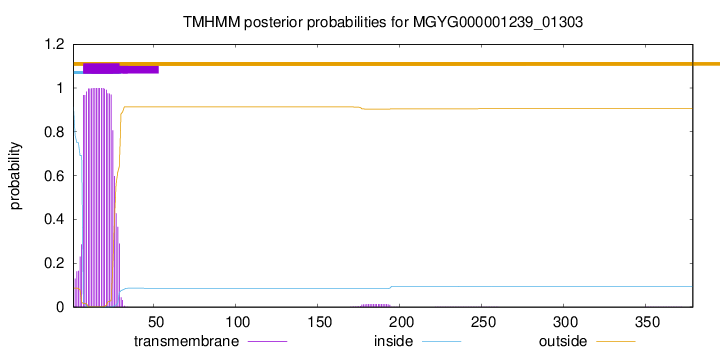
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.913201
0.084538
0.000553
0.000265
0.000201
0.001278