You are browsing environment: HUMAN GUT
MGYG000001253_00519
Basic Information
help
Species
Paramuribaculum sp001689535
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Paramuribaculum; Paramuribaculum sp001689535
CAZyme ID
MGYG000001253_00519
CAZy Family
GH95
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001253
2271514
MAG
Austria
Europe
Gene Location
Start: 57200;
End: 59467
Strand: -
No EC number prediction in MGYG000001253_00519.
Family
Start
End
Evalue
family coverage
GH95
57
715
3.4e-59
0.9598337950138505
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam18961
DUF5703_N
8.96e-04
277
291
273
287
Domain of unknown function (DUF5703). This is an N-terminal domain of unknown function mostly found in bacteria. It is possible that this domain might be a putative glycoside hydrolase. This family belongs to the Galactose Mutarotase-like superfamily.
more
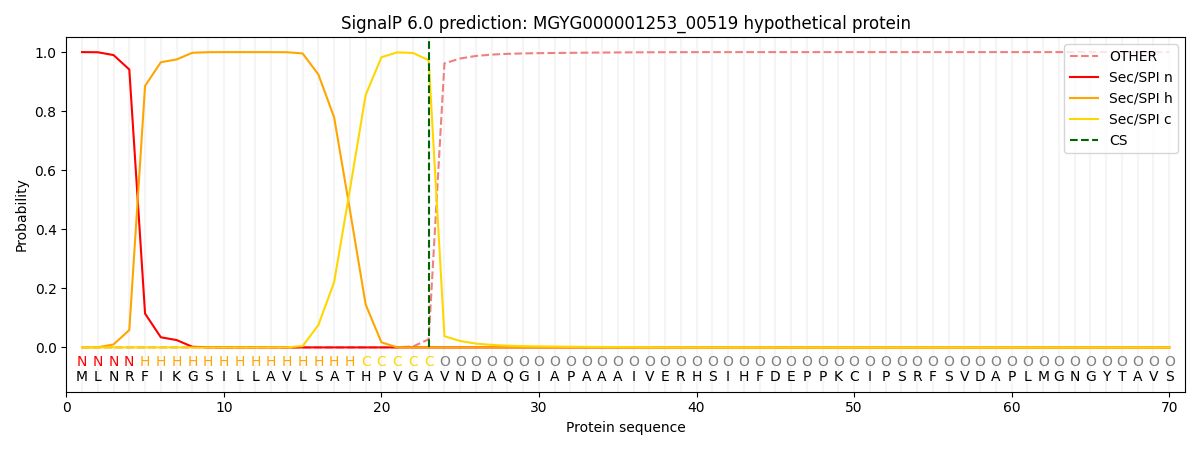
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000329
0.998932
0.000206
0.000191
0.000173
0.000149
There is no transmembrane helices in MGYG000001253_00519.