You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001309_03706
You are here: Home > Sequence: MGYG000001309_03706
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_F botulinum_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_F; Clostridium_F botulinum_A | |||||||||||
| CAZyme ID | MGYG000001309_03706 | |||||||||||
| CAZy Family | GH14 | |||||||||||
| CAZyme Description | Thermophilic beta-amylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2597696; End: 2599324 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH14 | 44 | 430 | 1.3e-136 | 0.9805825242718447 |
| CBM20 | 449 | 536 | 4e-18 | 0.9666666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01373 | Glyco_hydro_14 | 0.0 | 44 | 440 | 1 | 402 | Glycosyl hydrolase family 14. This family are beta amylases. |
| PLN02801 | PLN02801 | 2.27e-68 | 44 | 440 | 19 | 420 | beta-amylase |
| PLN02803 | PLN02803 | 3.95e-56 | 44 | 398 | 89 | 463 | beta-amylase |
| PLN02905 | PLN02905 | 1.61e-49 | 44 | 423 | 268 | 672 | beta-amylase |
| PLN00197 | PLN00197 | 1.21e-48 | 44 | 399 | 109 | 485 | beta-amylase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVQ43142.1 | 0.0 | 1 | 542 | 1 | 542 |
| AVQ39732.1 | 0.0 | 1 | 542 | 1 | 542 |
| APH19682.1 | 0.0 | 1 | 542 | 1 | 542 |
| AUM90908.1 | 0.0 | 1 | 542 | 1 | 542 |
| AUN02789.1 | 0.0 | 1 | 542 | 1 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VOC_A | 4.83e-175 | 40 | 450 | 5 | 416 | Crystalstructure of the catalytic domain of beta-amylase from paenibacillus polymyxa [Paenibacillus polymyxa] |
| 1B90_A | 6.86e-161 | 37 | 542 | 7 | 516 | BacillusCereus Beta-Amylase Apo Form [Bacillus cereus],1B9Z_A Bacillus Cereus Beta-Amylase Complexed With Maltose [Bacillus cereus],1J0Y_A Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_B Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_C Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Y_D Beta-amylase from Bacillus cereus var. mycoides in complex with glucose [Bacillus cereus],1J0Z_A Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_B Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_C Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J0Z_D Beta-amylase from Bacillus cereus var. mycoides in complex with maltose [Bacillus cereus],1J10_A beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_B beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_C beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J10_D beta-amylase from Bacillus cereus var. mycoides in complex with GGX [Bacillus cereus],1J11_A beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_B beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_C beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J11_D beta-amylase from Bacillus cereus var. mycoides in complex with alpha-EPG [Bacillus cereus],1J12_A Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_B Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_C Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J12_D Beta-Amylase from Bacillus cereus var. mycoides in Complex with alpha-EBG [Bacillus cereus],1J18_A Crystal Structure of a Beta-Amylase from Bacillus cereus var. mycoides Cocrystallized with Maltose [Bacillus cereus],1VEM_A Crystal Structure Analysis of Bacillus Cereus Beta-Amylase at the optimum pH (6.5) [Bacillus cereus],5BCA_A Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_B Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_C Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus],5BCA_D Beta-Amylase From Bacillus Cereus Var. Mycoides [Bacillus cereus] |
| 1VEO_A | 2.76e-160 | 37 | 542 | 7 | 516 | ChainA, Beta-amylase [Bacillus cereus] |
| 1ITC_A | 5.53e-160 | 37 | 542 | 7 | 516 | ChainA, Beta-Amylase [Bacillus cereus] |
| 1VEN_A | 1.57e-159 | 37 | 542 | 7 | 516 | ChainA, Beta-amylase [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19584 | 1.58e-235 | 9 | 542 | 6 | 549 | Thermophilic beta-amylase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 PE=1 SV=1 |
| P06547 | 1.59e-176 | 1 | 440 | 1 | 442 | Beta-amylase OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P96513 | 2.75e-176 | 6 | 450 | 5 | 451 | Beta-amylase (Fragment) OS=Cytobacillus firmus OX=1399 PE=3 SV=1 |
| P21543 | 4.03e-167 | 6 | 539 | 5 | 541 | Beta/alpha-amylase OS=Paenibacillus polymyxa OX=1406 PE=1 SV=1 |
| P36924 | 1.01e-159 | 37 | 542 | 37 | 546 | Beta-amylase OS=Bacillus cereus OX=1396 GN=spoII PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
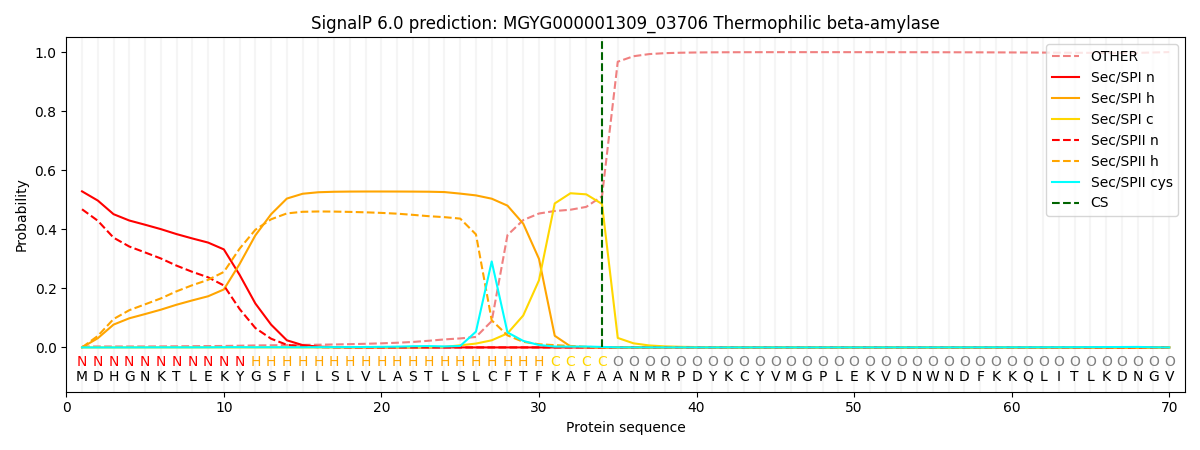
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004468 | 0.519253 | 0.474823 | 0.000858 | 0.000341 | 0.000239 |

