You are browsing environment: HUMAN GUT
MGYG000001313_02445
Basic Information
help
Species
Bacteroides eggerthii
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides eggerthii
CAZyme ID
MGYG000001313_02445
CAZy Family
GH5
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
659
73263.1
5.1751
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001313
4157999
Isolate
not provided
not provided
Gene Location
Start: 737098;
End: 739077
Strand: +
No EC number prediction in MGYG000001313_02445.
Family
Start
End
Evalue
family coverage
GH5
74
354
9e-112
0.9884169884169884
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam00150
Cellulase
9.88e-10
164
310
72
207
Cellulase (glycosyl hydrolase family 5).
more
pfam02368
Big_2
0.001
462
529
20
77
Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins.
more
NF033168
lipo_LIC10766
0.003
4
46
4
48
LIC_10766 family lipoprotein. Members of this family are lipoproteins found broadly in the genus Leptospira.
more
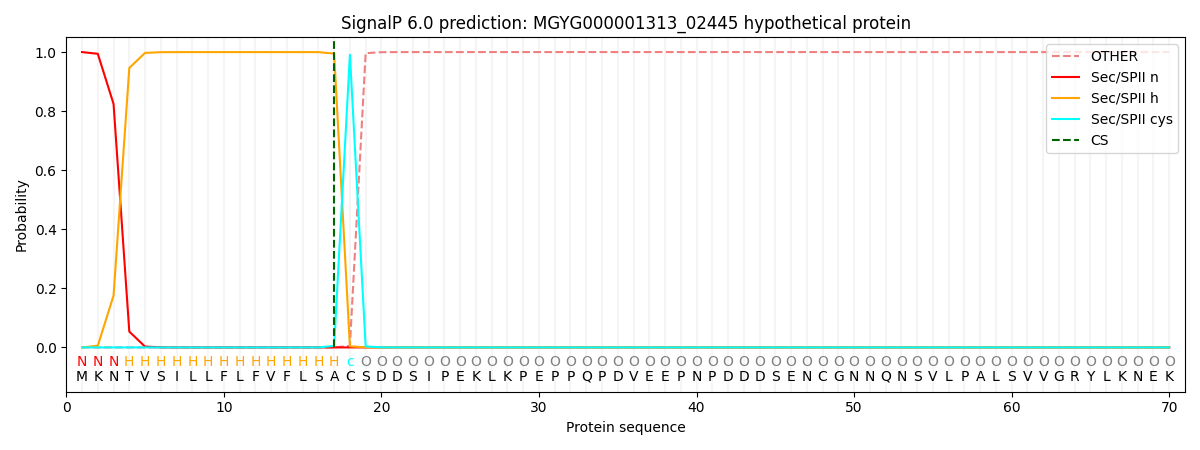
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000007
1.000073
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000001313_02445.