You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001346_01953
Basic Information
help
| Species |
Bacteroides uniformis
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides uniformis
|
| CAZyme ID |
MGYG000001346_01953
|
| CAZy Family |
GH30 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001346 |
4452540 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 645641;
End: 647542
Strand: +
|
No EC number prediction in MGYG000001346_01953.
| Family |
Start |
End |
Evalue |
family coverage |
| GH30 |
292 |
630 |
1.3e-16 |
0.7290167865707434 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG5520
|
XynC |
1.77e-10 |
293 |
632 |
108 |
429 |
O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| QNE42320.1
|
2.88e-177 |
23 |
632 |
2 |
642 |
| AYY14038.1
|
7.84e-09 |
304 |
633 |
167 |
518 |
| QTN00794.1
|
7.63e-06 |
297 |
438 |
150 |
293 |
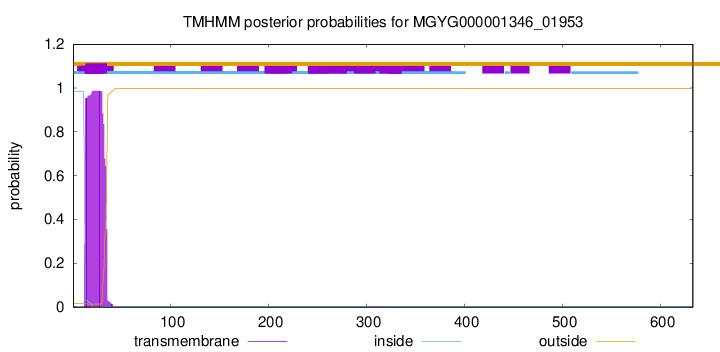
This protein is predicted as SP
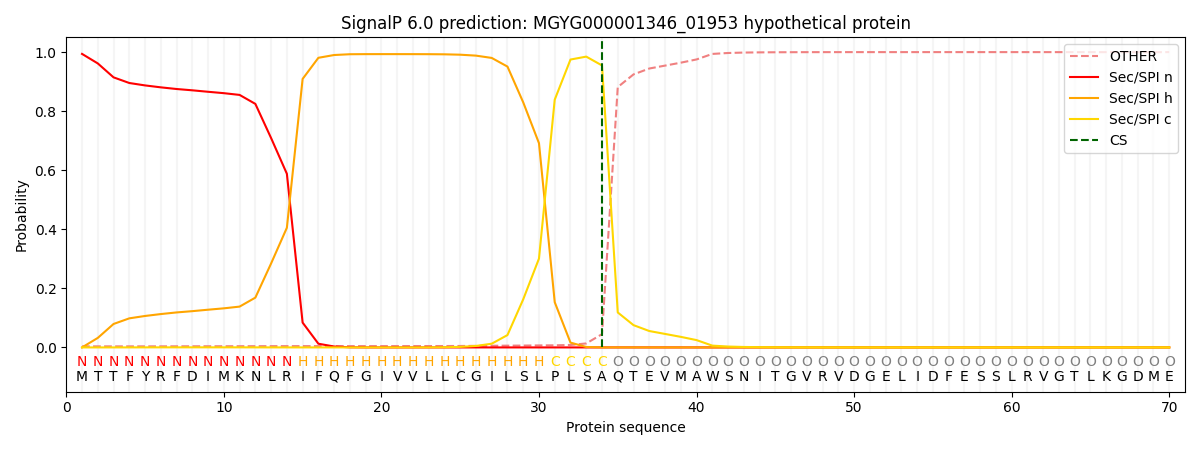
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.004713
|
0.991060
|
0.003540
|
0.000224
|
0.000211
|
0.000219
|