You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001377_01295
You are here: Home > Sequence: MGYG000001377_01295
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
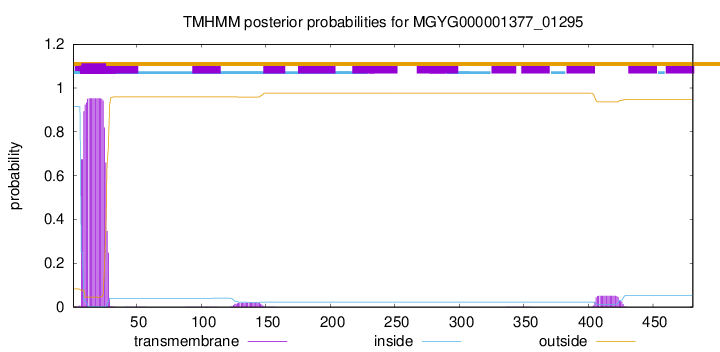
TMHMM annotations
Basic Information help
| Species | Dysgonomonas mossii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Dysgonomonas; Dysgonomonas mossii | |||||||||||
| CAZyme ID | MGYG000001377_01295 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 650994; End: 652439 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01825 | SGNH_hydrolase_peri1 | 2.55e-39 | 316 | 472 | 24 | 189 | SGNH_peri1; putative periplasmic member of the SGNH-family of hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| COG1388 | LysM | 3.87e-13 | 182 | 260 | 46 | 124 | LysM repeat [Cell wall/membrane/envelope biogenesis]. |
| pfam01476 | LysM | 1.18e-12 | 205 | 247 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| pfam13472 | Lipase_GDSL_2 | 2.08e-12 | 285 | 459 | 3 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 2.31e-11 | 285 | 466 | 5 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK56104.1 | 1.56e-135 | 1 | 475 | 1 | 484 |
| QIK61460.1 | 3.10e-134 | 26 | 475 | 2 | 469 |
| AWH83714.1 | 2.50e-124 | 37 | 477 | 41 | 469 |
| AOE53578.1 | 1.88e-122 | 40 | 476 | 48 | 472 |
| ALM49961.1 | 1.88e-122 | 40 | 476 | 48 | 472 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4K3U_A | 5.32e-10 | 50 | 470 | 38 | 369 | PeptidoglycanO-acetylesterase in action, 30 min [Neisseria meningitidis 4119],4K3U_B Peptidoglycan O-acetylesterase in action, 30 min [Neisseria meningitidis 4119],4K40_A Peptidoglycan O-acetylesterase in action, 0 min [Neisseria meningitidis LNP21362],4K40_B Peptidoglycan O-acetylesterase in action, 0 min [Neisseria meningitidis LNP21362],4K7J_A Peptidoglycan O-acetylesterase in action, 5 min [Neisseria meningitidis 4119],4K7J_B Peptidoglycan O-acetylesterase in action, 5 min [Neisseria meningitidis 4119] |
| 4K9S_A | 1.25e-09 | 50 | 148 | 38 | 131 | PeptidoglycanO-acetylesterase in action, setmet [Neisseria meningitidis 4119],4K9S_B Peptidoglycan O-acetylesterase in action, setmet [Neisseria meningitidis 4119] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
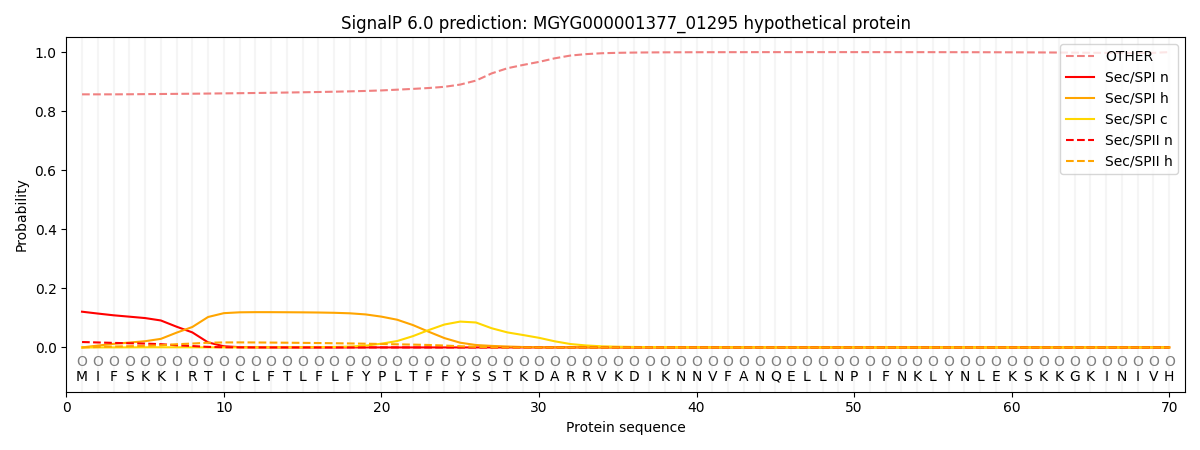
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.863592 | 0.112011 | 0.019075 | 0.000407 | 0.000299 | 0.004634 |