You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001380_00544
You are here: Home > Sequence: MGYG000001380_00544
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
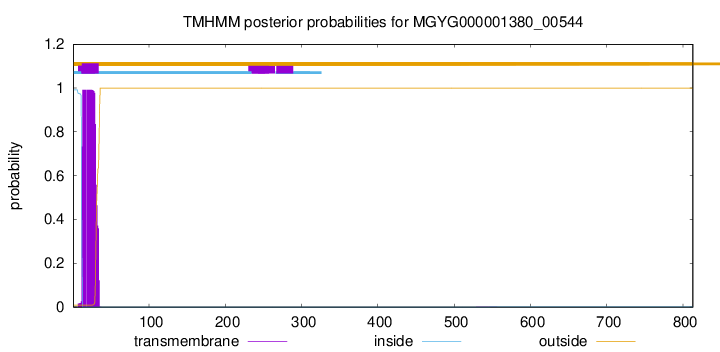
TMHMM annotations
Basic Information help
| Species | Ruminococcus_B gnavus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Ruminococcus_B; Ruminococcus_B gnavus | |||||||||||
| CAZyme ID | MGYG000001380_00544 | |||||||||||
| CAZy Family | GH98 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 147857; End: 150298 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 265 | 580 | 8e-43 | 0.981651376146789 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08306 | Glyco_hydro_98M | 6.42e-33 | 265 | 580 | 8 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| pfam08307 | Glyco_hydro_98C | 5.16e-26 | 583 | 755 | 1 | 174 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT30095.1 | 0.0 | 1 | 798 | 1 | 798 |
| QHB23852.1 | 0.0 | 1 | 798 | 10 | 807 |
| QEI31357.1 | 0.0 | 1 | 798 | 10 | 807 |
| AQM60602.1 | 5.63e-87 | 265 | 755 | 216 | 704 |
| BAB80035.1 | 1.91e-85 | 265 | 755 | 216 | 700 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7Q20_A | 0.0 | 39 | 798 | 1 | 760 | ChainA, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149],7Q20_G Chain G, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 7PMO_D | 0.0 | 39 | 798 | 1 | 760 | ChainD, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149],7PMO_G Chain G, Ruminococcus gnavus endogalactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 7Q1W_A | 0.0 | 40 | 798 | 1 | 759 | ChainA, Ruminococcus gnavus end galactosidase GH98 [[Ruminococcus] gnavus ATCC 29149] |
| 2WMI_A | 5.71e-84 | 260 | 755 | 23 | 512 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_A Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_B Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
| 2WMI_B | 7.95e-84 | 260 | 755 | 23 | 512 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6RUF5 | 4.22e-86 | 265 | 755 | 221 | 705 | Blood-group-substance endo-1,4-beta-galactosidase OS=Clostridium perfringens OX=1502 GN=eabC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
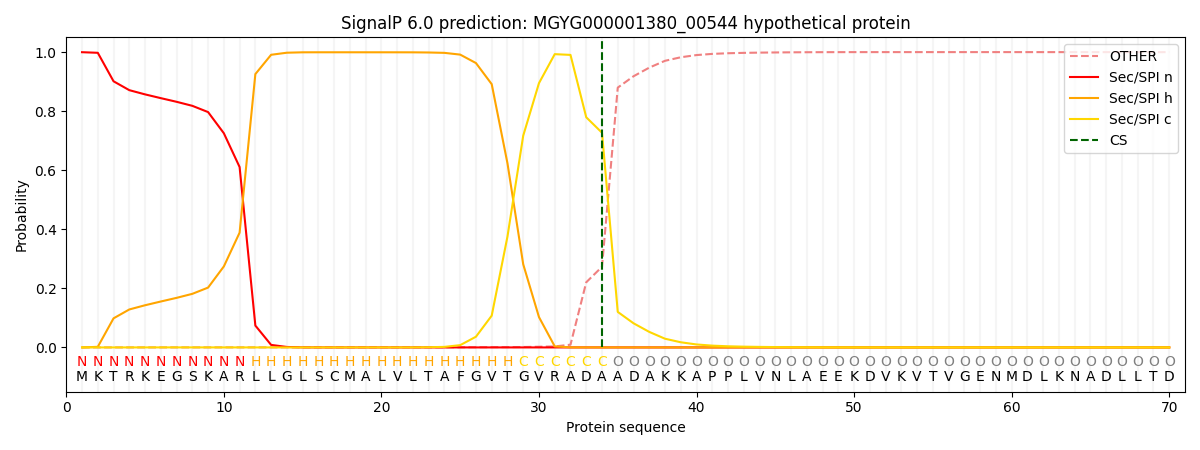
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000496 | 0.998237 | 0.000640 | 0.000270 | 0.000191 | 0.000152 |