You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001384_04678
You are here: Home > Sequence: MGYG000001384_04678
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ralstonia pickettii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Ralstonia; Ralstonia pickettii | |||||||||||
| CAZyme ID | MGYG000001384_04678 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Glucans biosynthesis glucosyltransferase H | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 130699; End: 133287 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 281 | 463 | 4.6e-19 | 0.9764705882352941 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2943 | MdoH | 0.0 | 111 | 860 | 3 | 732 | Membrane glycosyltransferase [Cell wall/membrane/envelope biogenesis, Carbohydrate transport and metabolism]. |
| PRK05454 | PRK05454 | 0.0 | 128 | 740 | 1 | 588 | glucans biosynthesis glucosyltransferase MdoH. |
| cd04191 | Glucan_BSP_MdoH | 1.47e-152 | 279 | 532 | 1 | 254 | Glucan_BSP_MdoH catalyzes the elongation of beta-1,2 polyglucose chains of glucan. Periplasmic Glucan Biosynthesis protein MdoH is a glucosyltransferase that catalyzes the elongation of beta-1,2 polyglucose chains of glucan, requiring a beta-glucoside as a primer and UDP-glucose as a substrate. Glucans are composed of 5 to 10 units of glucose forming a highly branched structure, where beta-1,2-linked glucose constitutes a linear backbone to which branches are attached by beta-1,6 linkages. In Escherichia coli, glucans are located in the periplasmic space, functioning as regulator of osmolarity. It is synthesized at a maximum when cells are grown in a medium with low osmolarity. It has been shown to span the cytoplasmic membrane. |
| cd06423 | CESA_like | 4.20e-16 | 281 | 469 | 1 | 174 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| pfam00535 | Glycos_transf_2 | 3.17e-14 | 281 | 461 | 2 | 164 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQK36403.1 | 0.0 | 34 | 862 | 1 | 829 |
| ANJ73769.1 | 0.0 | 1 | 859 | 1 | 859 |
| ANA33049.1 | 0.0 | 1 | 860 | 1 | 856 |
| ATG18811.1 | 0.0 | 1 | 860 | 1 | 856 |
| QIF08551.1 | 0.0 | 1 | 857 | 1 | 853 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46TZ4 | 0.0 | 33 | 858 | 17 | 841 | Glucans biosynthesis glucosyltransferase H OS=Cupriavidus pinatubonensis (strain JMP 134 / LMG 1197) OX=264198 GN=opgH PE=3 SV=1 |
| B7V3H0 | 0.0 | 38 | 848 | 14 | 819 | Glucans biosynthesis glucosyltransferase H OS=Pseudomonas aeruginosa (strain LESB58) OX=557722 GN=opgH PE=3 SV=1 |
| C3K816 | 0.0 | 38 | 849 | 14 | 821 | Glucans biosynthesis glucosyltransferase H OS=Pseudomonas fluorescens (strain SBW25) OX=216595 GN=opgH PE=3 SV=1 |
| Q1I3R6 | 0.0 | 38 | 849 | 14 | 819 | Glucans biosynthesis glucosyltransferase H OS=Pseudomonas entomophila (strain L48) OX=384676 GN=opgH PE=3 SV=1 |
| Q8XVC2 | 0.0 | 1 | 857 | 1 | 850 | Glucans biosynthesis glucosyltransferase H OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=opgH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.995954 | 0.004064 | 0.000009 | 0.000005 | 0.000003 | 0.000005 |
TMHMM Annotations download full data without filtering help
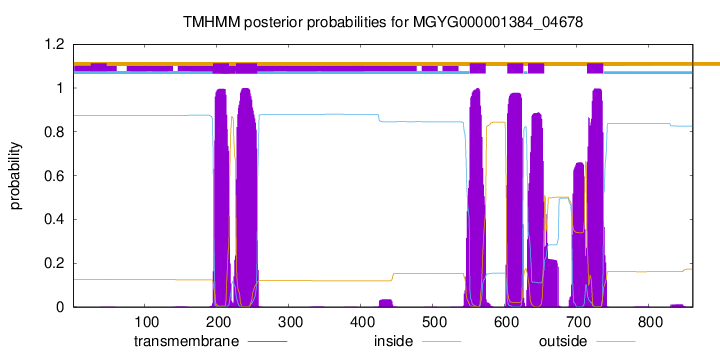
| start | end |
|---|---|
| 195 | 217 |
| 227 | 256 |
| 552 | 574 |
| 604 | 626 |
| 633 | 655 |
| 715 | 737 |
