You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001400_01869
You are here: Home > Sequence: MGYG000001400_01869
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
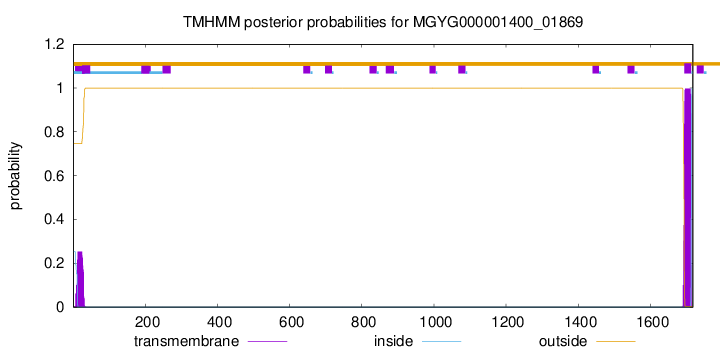
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium ramosum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium ramosum | |||||||||||
| CAZyme ID | MGYG000001400_01869 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | Hyaluronoglucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 226794; End: 231950 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 184 | 482 | 7.7e-98 | 0.9932203389830508 |
| CBM32 | 805 | 925 | 8.4e-17 | 0.8709677419354839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.45e-123 | 184 | 481 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 1.17e-23 | 37 | 177 | 1 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| cd00057 | FA58C | 9.27e-14 | 651 | 777 | 14 | 138 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 6.45e-13 | 650 | 779 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 2.79e-12 | 806 | 924 | 11 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQV04689.1 | 0.0 | 1 | 1718 | 1 | 1718 |
| QQY28342.1 | 0.0 | 1 | 1718 | 1 | 1718 |
| QPS12841.1 | 0.0 | 1 | 1718 | 1 | 1718 |
| QMW73892.1 | 0.0 | 1 | 1718 | 1 | 1718 |
| AYE33607.1 | 0.0 | 7 | 1629 | 6 | 1631 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PWI_A | 3.62e-173 | 25 | 628 | 21 | 620 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 6PV4_A | 3.28e-143 | 35 | 640 | 29 | 652 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PV5_A | 1.06e-66 | 40 | 554 | 42 | 554 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 5MI4_A | 1.13e-65 | 132 | 517 | 92 | 468 | BtGH84mutant with covalent modification by MA3 [Bacteroides thetaiotaomicron VPI-5482],5MI5_A BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482],5MI6_A BtGH84 mutant with covalent modification by MA3 in complex with Thiamet G [Bacteroides thetaiotaomicron VPI-5482],5MI7_A BtGH84 mutant with covalent modification by MA4 in complex with PUGNAc [Bacteroides thetaiotaomicron VPI-5482] |
| 2J4G_A | 1.65e-65 | 132 | 517 | 80 | 456 | Bacteroidesthetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482],2J4G_B Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 4.29e-272 | 35 | 1450 | 36 | 1484 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q89ZI2 | 1.33e-64 | 132 | 517 | 102 | 478 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q0TR53 | 2.39e-61 | 42 | 795 | 48 | 779 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 3.18e-61 | 42 | 759 | 48 | 736 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| O60502 | 7.01e-24 | 185 | 466 | 63 | 348 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
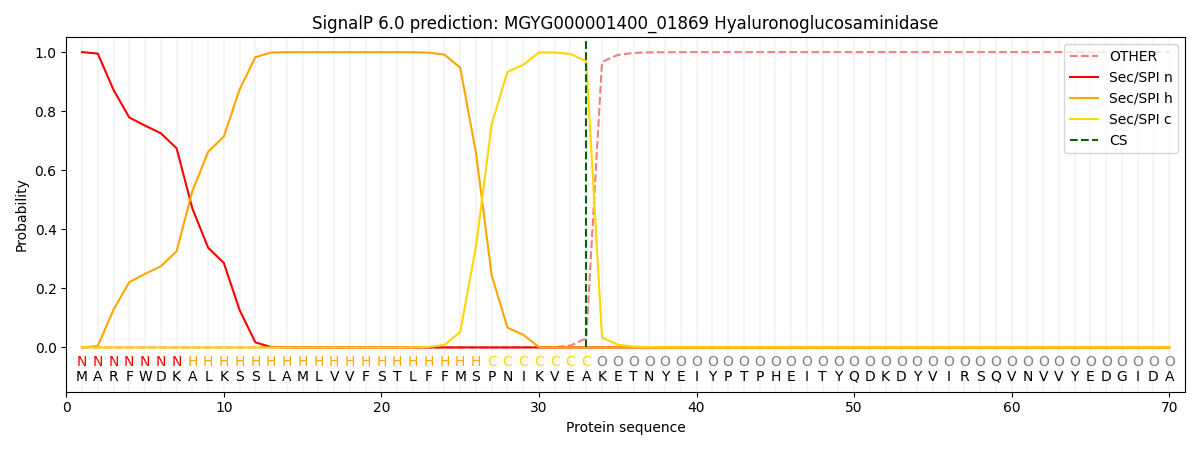
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999071 | 0.000175 | 0.000171 | 0.000149 | 0.000134 |