You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001411_00145
You are here: Home > Sequence: MGYG000001411_00145
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
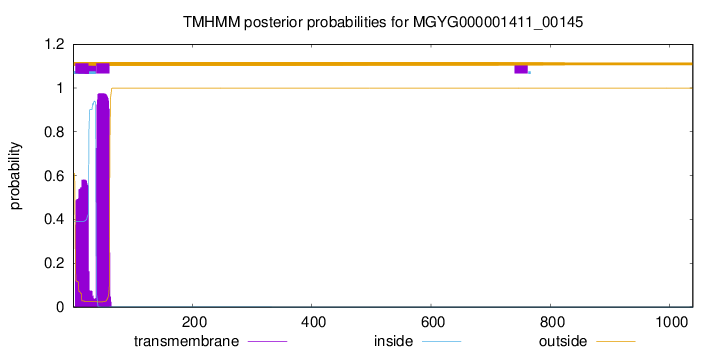
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp000308055 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp000308055 | |||||||||||
| CAZyme ID | MGYG000001411_00145 | |||||||||||
| CAZy Family | GH49 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 173337; End: 176456 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH49 | 92 | 693 | 4.9e-175 | 0.9872495446265938 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17433 | Glyco_hydro_49N | 3.08e-51 | 94 | 273 | 9 | 185 | Glycosyl hydrolase family 49 N-terminal Ig-like domain. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the N-terminal Ig-like fold. |
| pfam09985 | Glucodextran_C | 8.77e-35 | 789 | 1021 | 1 | 214 | C-terminal binding-module, SLH-like, of glucodextranase. Glucodextran_C is the C-terminal domain of glucodextranase-like proteins found in various prokaryotic membrane-anchored proteins. It shows homology to the carbohydrate-binding unit of some glycosidases. |
| cd09626 | DOMON_glucodextranase_like | 1.18e-33 | 789 | 1033 | 1 | 220 | DOMON-like domain of various glycoside hydrolases. This DOMON-like domain is found at the C-terminus of various bacterial proteins that play roles in metabolizing carbohydrates, such as glucodextranase (hydrolyzes alpha-1,6-glucosidic linkages of dextran from the non-reducing end), glucan alpha-1,4-glucosidase, pullulanase (degrades pullulan, a polysaccharide built from maltotriose units), arabinogalactan endo-1,4-beta-galactosidase, and others. Consequently, the DOMON-like domains in this family co-occur with catalytic domains from various glycosyl hydrolase families. The precise function of the DOMON domains in these proteins is not clear, they may be involved in interactions with carbohydrates. |
| pfam03718 | Glyco_hydro_49 | 1.96e-28 | 576 | 694 | 7 | 117 | Glycosyl hydrolase family 49. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the C-terminal pectate lyase like domain. |
| pfam18783 | IPU_b_solenoid | 9.85e-10 | 518 | 548 | 1 | 31 | Isopullulanase beta-solenoid repeat. IPU and dextranase repeat unit includes three (or one long and one short) parallel beta-strands. The repeat region as a whole folds into a beta-helix, known as beta-solenoid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CED92347.1 | 0.0 | 79 | 1033 | 56 | 987 |
| QPL04639.1 | 0.0 | 94 | 1037 | 65 | 981 |
| BAR04994.1 | 5.20e-299 | 94 | 1036 | 61 | 983 |
| BAR07052.1 | 3.32e-289 | 94 | 1033 | 74 | 1008 |
| QFU99755.1 | 1.20e-284 | 79 | 1037 | 52 | 979 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NZS_A | 9.18e-193 | 94 | 694 | 10 | 581 | DextranaseAoDex KQ11 [Pseudarthrobacter oxydans] |
| 3WWG_A | 1.19e-74 | 89 | 683 | 8 | 535 | Crystalstructure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_B Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_C Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_D Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger] |
| 1WMR_A | 8.08e-74 | 89 | 683 | 8 | 535 | CrystalStructure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1WMR_B Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_A Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_B Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],2Z8G_A Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger],2Z8G_B Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger] |
| 1OGM_X | 1.36e-72 | 96 | 692 | 15 | 571 | Dex49Afrom Penicillium minioluteum [Talaromyces minioluteus],1OGO_X Dex49A from Penicillium minioluteum complex with isomaltose [Talaromyces minioluteus] |
| 1UG9_A | 1.46e-59 | 696 | 1034 | 684 | 1017 | CrystalStructure of Glucodextranase from Arthrobacter globiformis I42 [Arthrobacter globiformis],1ULV_A Crystal Structure of Glucodextranase Complexed with Acarbose [Arthrobacter globiformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70744 | 9.92e-199 | 72 | 694 | 28 | 635 | Dextranase OS=Arthrobacter globiformis OX=1665 PE=3 SV=1 |
| P39652 | 4.23e-191 | 68 | 689 | 24 | 630 | Dextranase OS=Arthrobacter sp. (strain CB-8) OX=74565 PE=1 SV=1 |
| O00105 | 6.35e-73 | 89 | 683 | 23 | 550 | Isopullulanase OS=Aspergillus niger OX=5061 GN=ipuA PE=1 SV=1 |
| P48845 | 6.80e-73 | 96 | 692 | 49 | 605 | Dextranase OS=Talaromyces minioluteus OX=28574 GN=DEX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
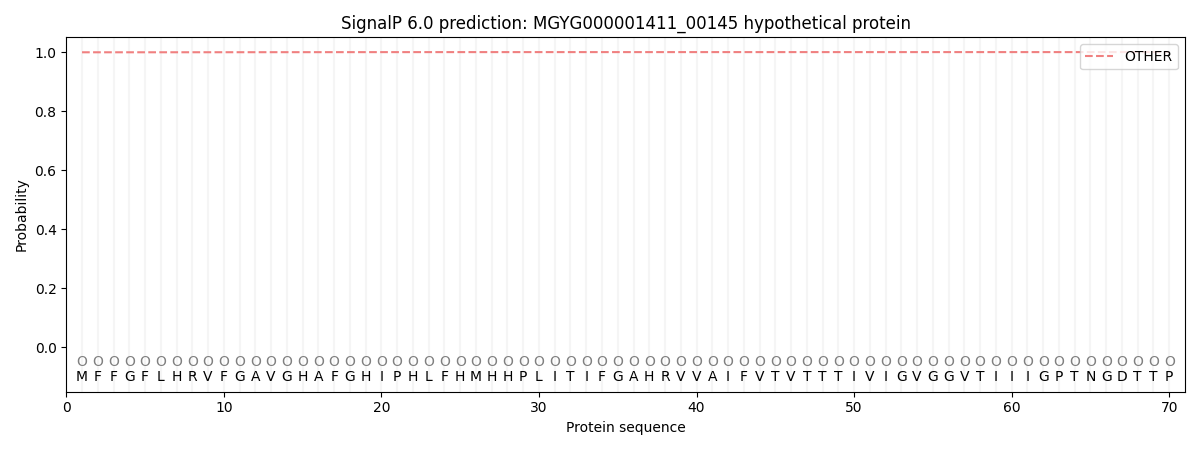
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999709 | 0.000298 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |