You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001414_01581
You are here: Home > Sequence: MGYG000001414_01581
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Peptoniphilus_E obesi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Peptoniphilaceae; Peptoniphilus_E; Peptoniphilus_E obesi | |||||||||||
| CAZyme ID | MGYG000001414_01581 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13552; End: 15063 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 298 | 438 | 9.2e-17 | 0.38943894389438943 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDZ74680.1 | 8.89e-184 | 1 | 501 | 3 | 507 |
| QXM05316.1 | 1.82e-100 | 36 | 503 | 42 | 509 |
| QOX62356.1 | 4.36e-99 | 20 | 503 | 33 | 513 |
| QZY55773.1 | 8.16e-97 | 36 | 497 | 60 | 517 |
| AOY76751.1 | 3.13e-96 | 41 | 501 | 69 | 520 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BVD_A | 5.38e-15 | 289 | 497 | 76 | 277 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 5.38e-15 | 289 | 497 | 76 | 277 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 5.89e-15 | 289 | 497 | 76 | 277 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2VI0_A | 9.66e-15 | 289 | 469 | 76 | 256 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 1.30e-14 | 289 | 497 | 76 | 277 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 2.07e-13 | 289 | 497 | 98 | 299 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
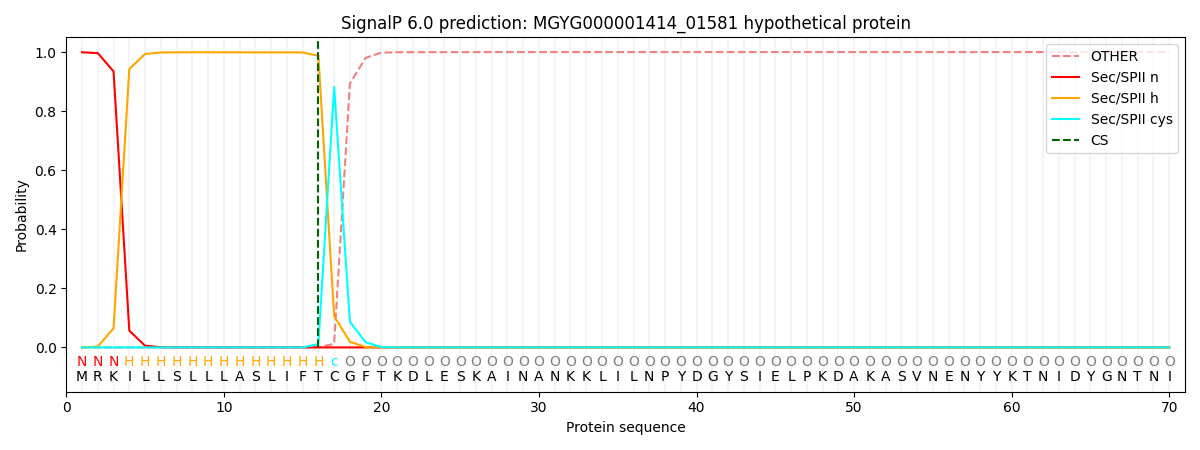
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000006 | 0.000565 | 0.999472 | 0.000000 | 0.000000 | 0.000000 |
